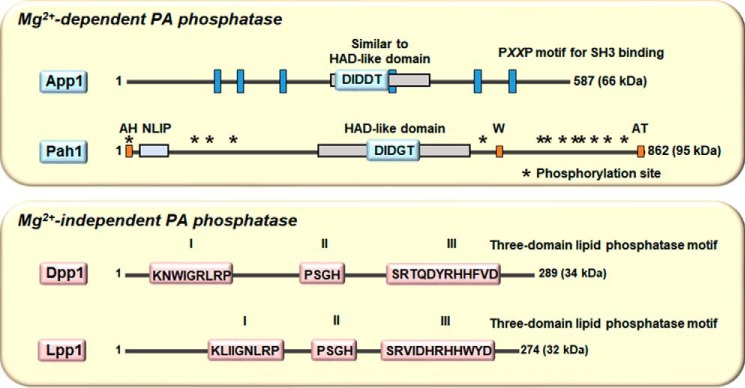
Figure 2.
Mg2+-dependent and -independent PA phosphatase enzymes in yeast have distinct catalytic motifs. The diagrams show linear representations of App1 and Pah1 (upper panel) and Dpp1 and Lpp1 (lower panel). The Mg2+-dependent PA phosphatase activities of App1 and Pah1 are governed by the DXDX(T/V) motifs in the HAD-like domain, whereas the Mg2+-independent PA phosphatase activities of Dpp1 and Lpp1 are governed by the three-domain phosphatase motif. For Pah1, the approximate positions are indicated for the amphipathic helix (AH) required for ER membrane interaction (77), the NLIP and HAD-like domains that are required for PA phosphatase activity (116), the acidic tail (AT) required for interaction with Spo7 of the Pah1 phosphatase (70, 77, 80), the tryptophan (W) residue within the C-terminal conserved sequence WRDPLVDID required for Pah1 function (117), and the sites phosphorylated by Pho85–Pho80, Cdc28–cyclin B, protein kinase A, protein kinase C, and casein kinase II that regulate the location, activity, and proteasomal degradation of Pah1 (76, 78, 79, 83, 84, 118).