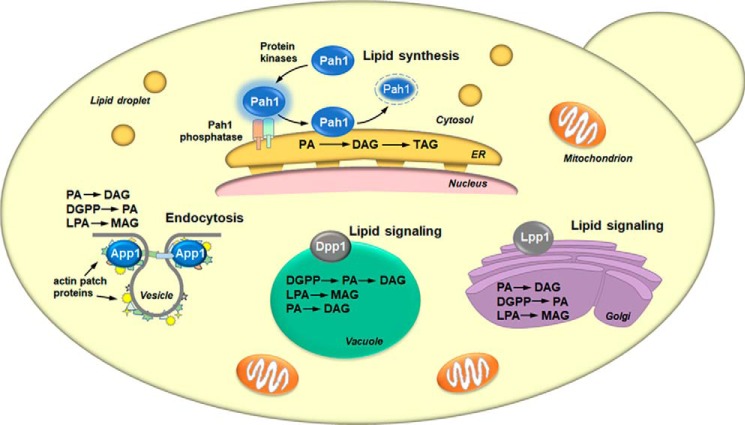
Figure 4.
Cellular locations and roles of yeast PA phosphatases. The figure depicts a yeast cell with the cellular locations of the App1, Dpp1, Lpp1, and Pah1 PA phosphatase enzymes and their physiological roles. The lipid phosphate phosphatase reaction catalyzed by each enzyme is indicated in the order of substrate preference. For Pah1, the enzyme in the cytosol is phosphorylated by multiple protein kinases. The phosphorylated Pah1 (indicated by the blue halo) translocates to the ER membrane through its dephosphorylation by the Pah1 phosphatase, which is composed of Nem1 (catalytic subunit, green) and Spo7 (regulatory subunit, peach). Dephosphorylated Pah1 that is associated with the ER membrane catalyzes the conversion of PA to DAG, which is then acylated to form TAG. Dephosphorylated Pah1 is degraded by the proteasome (indicated by the dashed lines). App1 associates with actin patch proteins at cortical actin patches. The App1 enzyme is postulated to be involved in the formation of endocytic vesicles. The vesicle diagram is adapted from Ref. 74. Dpp1 and Lpp1, respectively, are integral membrane proteins in the vacuole and Golgi apparatus. They are postulated to be involved with lipid signaling by controlling the amounts of bioactive lipids.