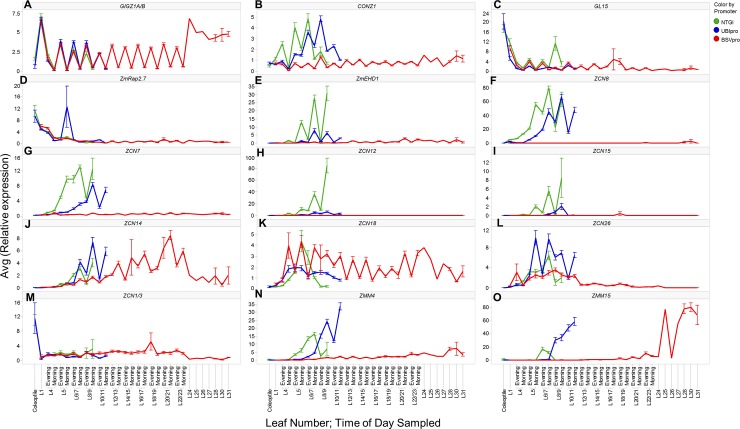
Fig 5. Expression patterns of flowering regulators in leaves of non-transgenic (NTG), UBIpro:ZmCCT10, and BSVpro:ZmCCT10 transgenic plants.
A) GIGZ1A/1B, a circadian clock gene. B) CONZ1, a circadian clock gene. C) GL15, the regulator of transition from a juvenile to adult leaf identity, a target of miR172. D) ZmRap2.7, a repressor of flowering, a target of miR172. E) ZmEHD1, an activator of flowering. F) ZCN8, the major florigen gene, an activator of flowering. G) ZCN7, another putative florigen gene, an activator of flowering. H) ZCN12, I) ZCN15, J) ZCN14, K) ZCN18, FT-like genes with florigenic activity in Arabidopsis. L) ZCN26, a FT-like gene with no florigenic activity in Arabidopsis. M) ZCN1/3, TFL1-like genes, repressors of flowering. N) ZMM4 and O) ZMM15 MADS genes, activators of flowering, markers for floral transition. The X-axis represents the tissue sampled: Coleoptile, leaves 1 to 22 (sampled either in the morning or evening), leaves 24 to 31 (sampled in the evening). The Y-axis represents the average relative gene expression normalized against eukaryotic initiation factor 4-gamma (GenBank EU967723). Error bars represent ± the SE.