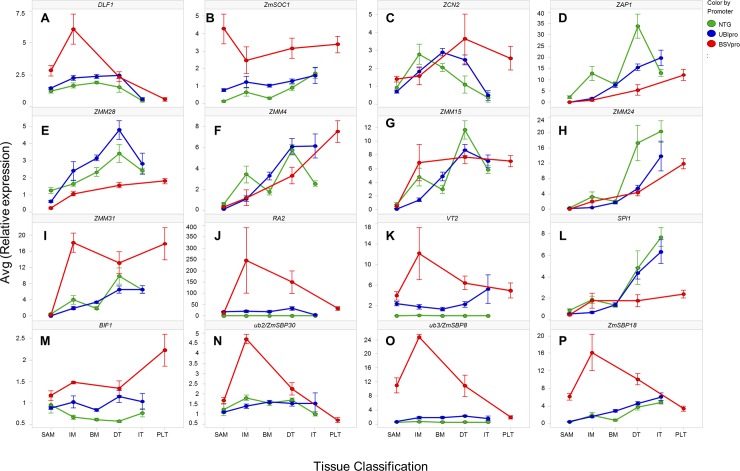
Fig 6. Expression patterns of inflorescence developmental genes in the shoot apices of non-transgenic (NTG), UBIpro:ZmCCT10, and BSVpro:ZmCCT10 transgenic plants.
A) DLF1 and B) ZmSOC1, floral transition genes. C) ZCN2, maize homolog of Arabidopsis TFL1, a repressor of the floral transition. AP1-like MADS genes D) ZAP1, E) ZMM28, F) ZMM4, and G) ZMM15. SEP-like MADS genes H) ZMM24 and I) ZMM31. J) RA2, SPM determinacy gene. K) VT2 and L) SPI1, auxin biosynthesis genes. M) BIF1, auxin signaling gene. N) UB2/SBP30. O) UB3/ZmSBP8, and P) ZmSBP18 genes, targets of miR156. The X-axis represents developmental stages of apices as defined in Fig 5. SAM: shoot apical meristem, IF: inflorescence meristem, BM: branch meristem, DT: developing tassel, IT: immature tassel, PLT: plantlets. The Y-axis represents the average relative gene expression normalized against eukaryotic initiation factor 4-gamma (GenBank EU967723). Error bars represent ± the SE.