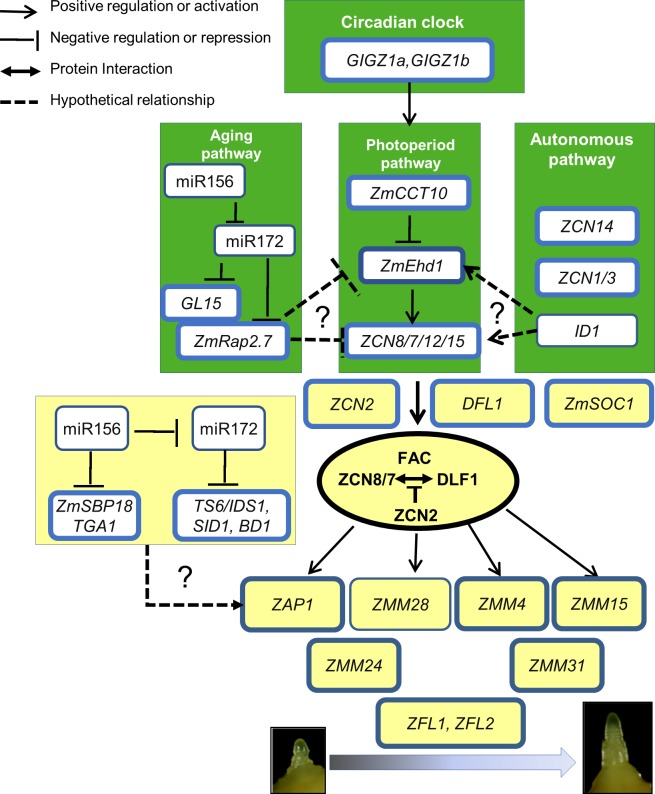
Fig 7. A conceptual gene network model for regulation of flowering time in maize.
The model is divided into genes expressed in leaves (green background) and those expressed in the shoot apical meristem (yellow background). Genes tested in this study are framed by a blue outline. In the leaves, three pathways are depicted: aging, photoperiod and autonomous. The photoperiod pathway is represented by the module ZmCCT10 (rice Ghd7)—ZmEhd1–ZCN8/7/12/15 (FT-like genes) which is conserved in other short-day monocots, rice and sorghum. ZmCCT10 is likely a repressor of the flowering activator ZmEhd1 gene which is a homolog of rice Ehd1 and sorghum SbEhd1. ZmCCT10 is not expressed under inductive SDs releasing ZmEhd1 from repression and activating the FT-like ZCN8/7/12/15 genes. ZmCCT10 is expressed under non-inductive LDs leading to repression of the FT-like ZCN8/7/12/15genes which occurs directly or indirectly via repression of ZmEhd1. However, the FT-like ZCN14 gene is not repressed by ZmCCT10 and may be placed in the autonomous pathway. In the aging pathway, ZmRap2.7 represses flowering via suppression of ZmEhd1 or ZCN8/7 genes. In the autonomous pathway, ID1 is required for activation of ZCN8/7 and possibly ZmEhd1 or other upstream regulators of ZCN8/7. Once transported to the shoot apical meristem (SAM), the florigenic ZCN8/7proteins interact with DLF1 forming the florigen activation complex (FAC, yellow oval). The florigen-antagonistic protein ZCN2 may compete with ZCN8/7 in the FAC leading to delayed flowering. Expression of DFL1 and ZCN2 is independent of the photoperiod pathway. Formation of the FAC in the SAM induces transcription of the AP1-like MADS box meristem identity genes ZAP1, ZMM4 and ZMM15 marking the onset of inflorescence specification. The ZFL1/2 genes are positioned downstream of the AP1-like genes.