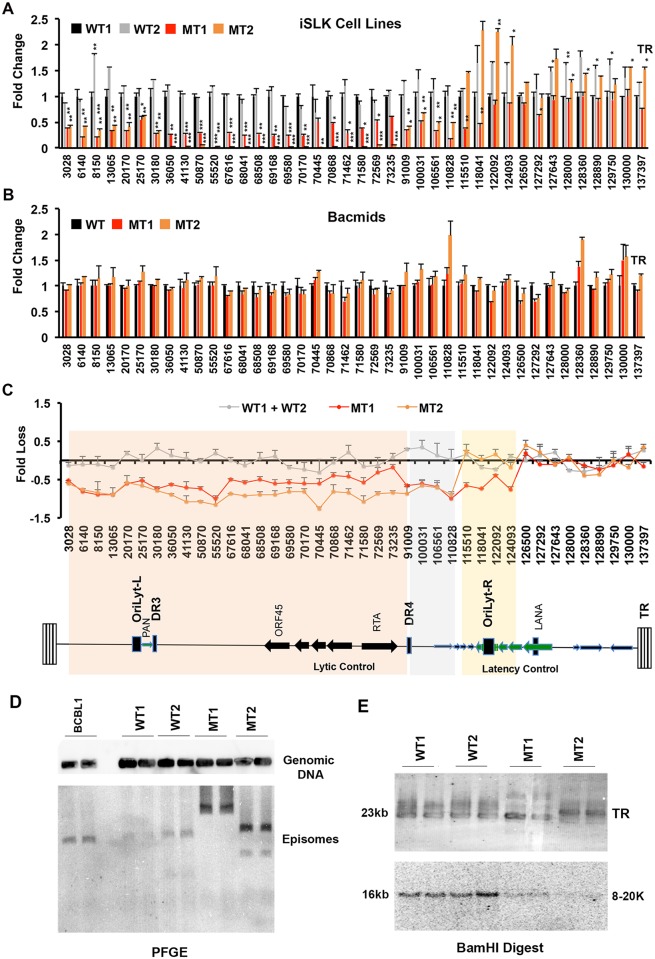
Fig 7. LANA oligomerization is important for viral genome integrity.
(A) RFP-LANA WT1, WT2, MT1, and MT2 stable iSLK cell lines were analyzed by qPCR for copy number variation using primers spanning KSHV genome, as indicated on X-axis. (B) RFP-LANA WT, MT1, and MT2 bacmid DNA was quantified by qPCR as in panel A. (C) Fold loss of KSHV DNA relative to bacmid control for MT1, MT2, and the average of WT1+WT2. Region of genetic instability are highlighted with salient viral genes and features indicated. * p value < .05, ** p value < .01 using student t-test. (D) BCBL1 or KSHV Bac RFP-LANA WT1, WT2, MT1, or MT2 were analyzed by agarose plug PFGE and Southern blot with KSHV bacmid probe (Episomes) or 18S DNA (Genomic DNA). Agarose plugs containing 106 cells were analyzed in duplicate. (E) Total genomic DNA from or KSHV Bac RFP-LANA WT1, WT2, MT1, or MT2 was digested with BamHI and assayed by PFGE and Southern blot with probes to TR (top) or KSHV genomic DNA between coordinates 2kb-18kb on the KSHV genome.