Figure 6. Substrate binding site and kinetics of ZmALDH12.
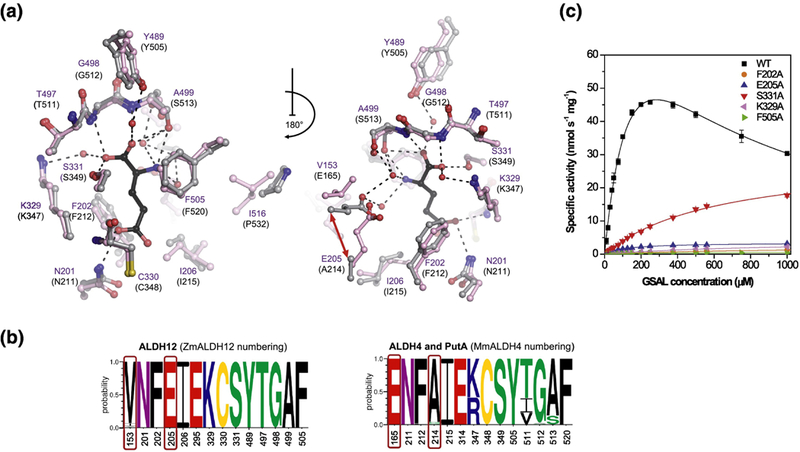
(a) The substrate cavity and surrounding residues in ZmALDH12 colored in pink (PDB ID: 6D97, this work). Residues of MmALDH4 (gray, PDB ID: 3V9K)35 involved in binding of the reaction product glutamate (colored in black) are shown for comparison. A repositioning of active-site glutamate E165 in MmALDH4 and E205 in ZmALDH12 is indicated by the red arrow. (b) An overview of the conservation of amino acid residues forming the substrate-binding site in enzymes from the ALDH12 family, which is compared with those found in ALDH4 used as a reference and numbered as MmALDH4. Sequence logos were made using WebLogo 3. (c) Saturation curves for active-site variants of ZmALDH12 with GSAL. The data were measured in 100 mM pyrophosphate buffer, pH 7.5 using 3.0 mM NAD+ as a coenzyme. Error bars stand for S.D. from four measurements.
