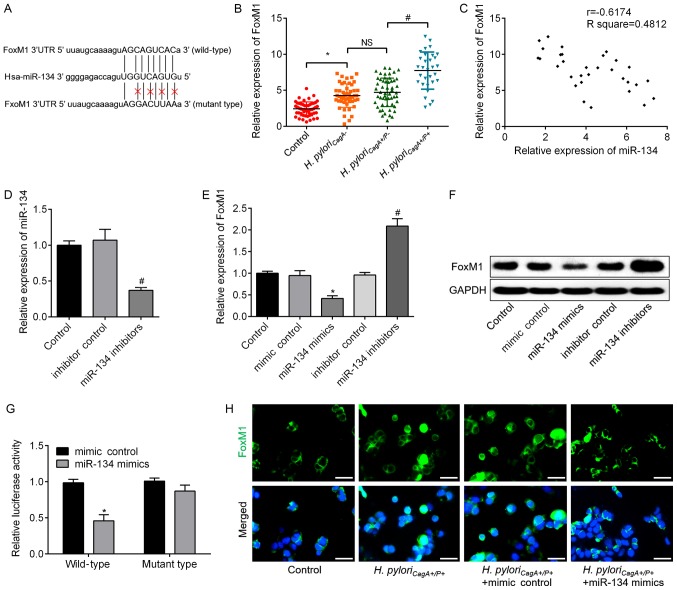
Figure 4.
FoxM1 is a downstream target of miR-134, which directly binds to the 3′UTR of FoxM1. (A) Putative miR-134-binding sites in the 3′UTR of FoxM1 mRNA. A mutation was generated in the FoxM1 3′UTR sequence in the complementary site for the seed region of miR-134. (B) Relative expression levels of FoxM1 in GC tissues without H. pylori infection, and tissues infected with H. pyloriCagA−, H. pyloriCagA+/P− and H. pyloriCagA+/P+. (C) A negative correlation was observed between FoxM1 mRNA and miR-134 expression in GC tissues infected with H. pyloriCagA+/P+. (D) Expression levels of miR-134 in SGC-7901 cells transfected with inhibitor control or miR-134 inhibitors, as determined by RT-qPCR analysis. (E) mRNA expression levels of FoxM1 were analyzed by RT-qPCR analysis in SGC-7901 cells transfected with mimic control, miR-134 mimics, inhibitor control or miR-134 inhibitors. GAPDH was used as an internal control. (F) Protein expression levels of FoxM1 were determined by western blotting in the groups. GAPDH was used as an internal control. (G) Luciferase assay in 293T cells co-transfected with miR-134 mimics or mimic control and a luciferase reporter containing wild type or mutant FoxM1 3′UTR. (H) Immunofluorescence analysis revealed the alteration of FoxM1 in H. pyloriCagA+/P+-infected SGC-7901 cells transfected with miR-134 mimics or mimic control. Images merged with DAPI are presented. Scale bar, 50 µm. *P<0.05 vs. the mimic control group; #P<0.05 vs. the inhibitor control group. 3′UTR, 3′-untranslated region; CagA, cytotoxin-associated gene A; H. pyloriCagA-, CagA-negative H. pylori; H. pyloriCagA+/P−, CagA-positive and PBP1A mutation-negative H. pylori; H. pyloriCagA+/P+, CagA- and PBP1A mutation-positive H. pylori; FoxM1, Forkhead box M1; H. pylori, Helicobacter pylori; miR-134, microRNA-134; PBP1A, penicillin-binding protein 1A; RT-qPCR, reverse transcription-quantitative polymerase chain reaction.