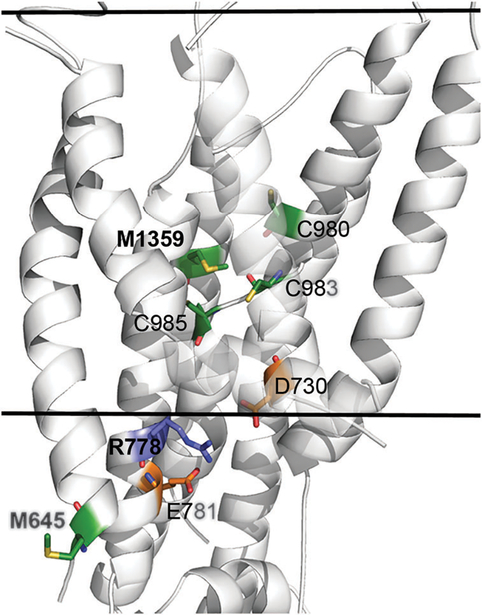
Fig. 3. Structural location and molecular interactions of M645, R778 and M1359.
The model-structure of ATP7B is shown as white cartoon, and viewed from the side with the cytoplasm below. The view is focused on the membrane domain, and the estimated membrane boundaries, calculated by the PPM webserver62 (https://opm.phar.umich.edu/ppm_server.php), are marked by the two solid lines. M645, R778, M1359 and interacting residues of interest are shown as sticks, with Met and Cys in green, Arg in blue and Asp and Glu in orange. Nitrogen, Oxygen and Sulfur atoms of these residues are colored blue, red and yellow, respectively. For clarity, the C-termini of TM2 and TM3 are partly transparent. The variable position M645 is situated at the beginning of TM1, facing outwards in contrast to highly conserved M1359 that is situated at the protein core, facing several other potential highly conserved copper ligands (sticks). The conserved R778 is situated at the region corresponding to the suggested copper entry site of LCopA, in close proximity to highly conserved acidic residues, potentially forming stabilizing interactions.