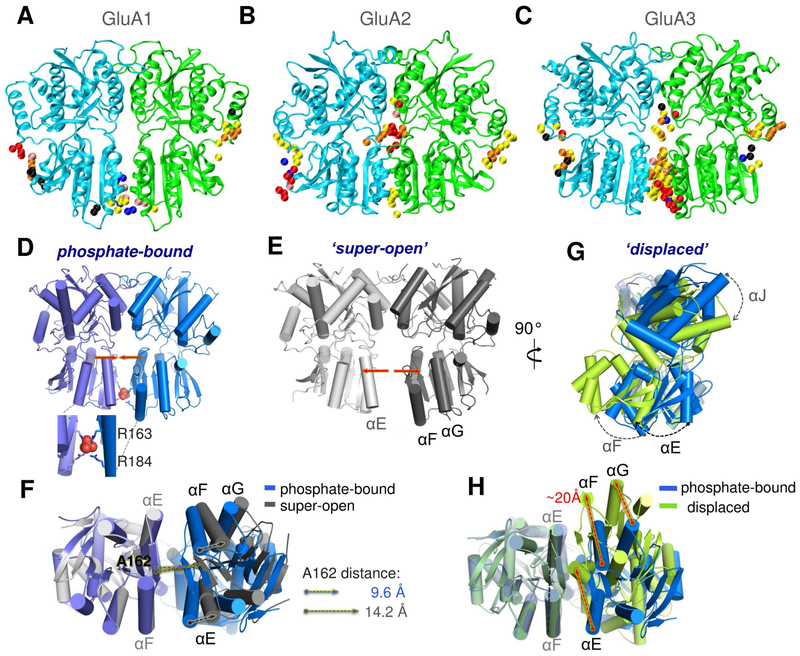
Figure 4. Distinctive ligand binding and conformational flexibility of GluA3 NTD dimers revealed by druggability simulations and X-ray crystallography.
(A-C) Conserved probe binding patterns observed in multiple druggability simulations of GluA1 (A), GluA2 (B) and GluA3 (C) NTD dimers. In contrast to GluA1 and GluA2, GluA3 features a high affinity ligand-binding site at the LL interface (see contact distributions in Figure S4). (D-H) Our crystal structures of the GluA3 NTD reveal unique dimeric states: one with phosphate bound between the pairs of R163 and R184 residues (D), which is comparable to the previously resolved open conformer (PDB ID: 3O21 dimer CD; see Figure S5); a super-open form (E); and a displaced dimer found in the same crystal as the phosphate bound form (G). Bottom views in F and H compare the PO43−-bound structure to the super-open and displaced structures, respectively. Phosphate binding is mimicked by dianionic methylphosphate binding in additional druggability simulations (see Figure S7).