Figure 2. Samples for cryoEM studies and resolution of the cryoEM maps.
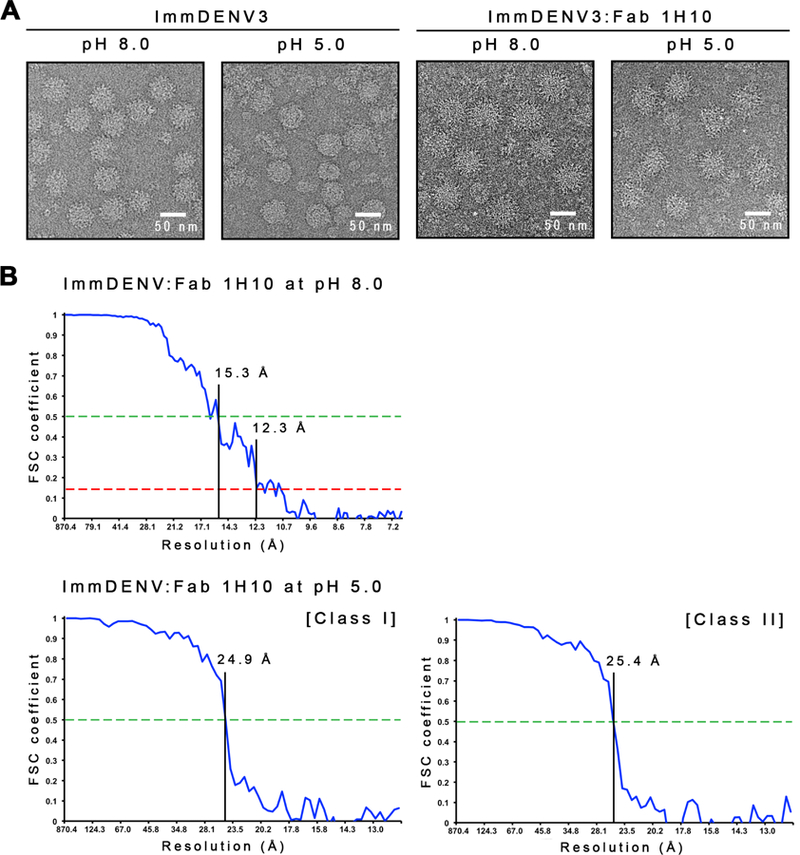
(A) Micrographs of the uncomplexed immDENV3 and the immDENV3:Fab 1H10 complexes at pH 8.0 and 5.0. The uncomplexed immDENV3 at pH 8.0 micrograph showed the sample contains mostly spiky fully immature virus. At pH 5.0, the uncomplexed virus becomes unstable and the surface of particles looked smoother. In the immDENV3:Fab 1H10 complex samples, at both pH 8.0 and 5.0, particles appear to have a bigger diameter and spikier, indicating the binding of Fab molecules.
(B) [Top] Fourier Shell Correlation (FSC) curve of immDENV:Fab 1H10 at pH 8.0 cryoEM map determined by gold-standard FSC procedure. The 12 Å resolution of the map is estimated by using FSC 0.143 correlation cut-off. [Bottom] Fourier shell correlation curve of class I and II immDENV:Fab 1H10 at pH 5.0 cryoEM maps. The FSC curve was plotted from two reconstructed maps of two half-datasets of the final iteration step and the resolution was estimated by using 0.5 cut-off. Class I and II pH 5.0 complexed structures were determined to a resolution of ~25Å, suggesting the structures were flexible.
See also Figure S1.
