Abstract
Incorporation of a MYC immunohistochemical stain in the work-up of large B-cell lymphomas has become common in hematopathology practice. Evaluation of this stain can be difficult due to staining heterogeneity and can have inter-observer variability, particularly when performed on entire tumor sections. We identified 87 cases of aggressive B cell lymphoma (34 core needle and 53 excisional biopsies) and compared the following methods of MYC immunohistochemical staining evaluation: the original pathologist’s interpretation, a systematic retrospective method of evaluation by manual analysis, and a retrospective method of evaluation by digital image analysis (using scanned slides analyzed via the Aperio Nuclear algorithm). Overall, concordance among these methods was around 80% with kappa statistics showing good agreement. However, nearly one-third of our cases had a percent MYC positivity in the 30% to 50% range and, for these cases, concordance among the various methods was marginal/poor. This suggests limited utility as a prognostic or predictive marker using 40% as a cut-off value. In our series, core biopsy specimens were poor predictors of MYC gene rearrangement and there was no association between MYC immunohistochemical stain and MYC gene gain/amplification. Our retrospective digital image analysis showed strong correlation in MYC percent positivity with our retrospective manual review (correlation coefficient of 0.90) and similar concordance to pathologist interpretation as among pathologists, suggesting digital image analysis is a viable alternative to manual determination of MYC percent positivity. Digital image analysis provides further opportunities for more sophisticated and standardized scoring systems, which may be helpful in future prognostic/predictive studies.
Keywords: MYC, diffuse large B-cell lymphoma, DLBCL, high-grade B-cell lymphoma, digital image analysis, prognosis, immunohistochemistry
1. Introduction
Rearrangement of the MYC gene is well described in aggressive B-cell lymphomas including Burkitt lymphoma (nearly all cases) and diffuse large B-cell lymphoma (8-14% of cases)(1). A higher proportion of diffuse large B-cell lymphomas (DLBCL) show MYC protein expression (detected in approximately 30% of cases) than MYC gene rearrangement (2, 3). Translocation of the BCL2 gene occurs in 20-30% of DLBCL cases and rearrangement of the 3q27 region involving BCL6 is seen in up to 30% of DLBCL (1). Cases of DLBCL or cases with morphologic features of both Burkitt lymphoma and DLBCL (referred to in the 2008 World Health Organization classification as “B-cell lymphoma, unclassifiable, with features intermediate between DLBCL and Burkitt lymphoma”) and harboring a MYC (8q24) rearrangement in combination with a BCL2 (18q21) and/or a BCL6 (3q27) rearrangement have a relatively low complete response rate and short overall survival with R-CHOP or comparable therapies. These lymphomas, sometimes referred to as “double-hit” and “triple-hit” lymphomas, are separately classified according to the 2016 World Health Organization (WHO) classification as “high-grade B-cell lymphoma with MYC and BCL2 and/or BCL6 rearrangements.” Dual positivity by immunohistochemical stains for MYC and BCL2 in DLBCL (double-expression of MYC and BCL2 proteins) is associated with inferior survival in most studies (2-4). Such double-expression may be a prognostic marker but does not warrant separate classification according to the 2016 WHO classification.
As MYC and BCL2 protein expression may have prognostic significance and there is theoretical use for MYC immunohistochemical staining to triage which aggressive B-cell lymphomas require genetic studies to evaluate for double- or triple-hit, the incorporation of a MYC immunohistochemical stain has become common in hematopathology practice. After the standard validation process for a new immunohistochemical stain (5), our institution began offering the MYC immunohistochemical stain with a recommended cut-off (≥ 40% as positive) in early 2015. Unfortunately, immunohistochemical evaluation of this stain can have inter-observer variability, particularly when performed on entire tumor sections by a diverse group of pathologists (6). Digital image analysis is an emerging reproducible method of quantifying positivity by immunohistochemical stains (7-9).
We studied cases of aggressive B-cell lymphoma (Burkitt lymphoma, diffuse large B-cell lymphoma, and high-grade B-cell lymphoma) with the MYC immunohistochemical stain performed as part of routine clinical care. Our aims were (1) to compare multiple methods of MYC immunohistochemical staining evaluation (the original pathologist’s interpretation, a systematic retrospective method of evaluation by manual analysis, and a systematic retrospective method of evaluation by digital image analysis) and (2) to assess the ability of MYC immunohistochemical staining to predict a MYC gene rearrangement or MYC gene gain/amplification.
2. Materials and Methods
Our institutional review board approved this retrospective study. We used a Sunquest CoPathPlus (version 6.1.1) database search to identify cases of large B-cell lymphoma with a MYC immunohistochemical stain performed as part of routine clinical care from the time the stain was made available in March 2015 through December 31, 2016. During this time period, the MYC immunohistochemical stain was not used as a triaging tool for cytogenetic analysis at our institution. The MYC immunohistochemical stain (clone Y69; dilution 1:229; Abcam, Cambridge, MA, USA) was performed using the standard procedures of our immunohistochemistry laboratory at the time. The original pathologist’s interpretation of MYC staining (positive, negative, or equivocal) was obtained from pathology reports and is referred to as “original IHC interpretation” in the Results section. The results of cytogenetic studies (fluorescence in-situ hybridization, FISH), performed using standard methods for our laboratory at the time of biopsy, were recorded when performed on the same tissue sample as the immunohistochemical stain. A MYC breakapart probe was used (Vysis LSI MYC dual color break apart rearrangement probe, Abbott Molecular, Abbott Park, Illinois, USA) to determine MYC rearrangement and gain/amplification status. Gain/amplification of the MYC gene for the purposes of this study was defined as greater than 2 (>2) MYC fusion signals. We evaluated the hematoxylin-and-eosin (H&E) and MYC immunohistochemistry stained slides for sufficient tissue and quality of specimen/staining with any cases with insufficient tissue or missing/unavailable slides excluded. Up to ten areas on each H&E slide, measuring approximately 1 mm x 1 mm, were manually marked with a dotting pen. Areas with the best quality and most neoplasm-rich tissue were selected while areas with crush artifact, fibrosis, and necrosis were avoided. The corresponding areas on the MYC stained slides were also manually marked. Figure 1 illustrates the methods of retrospective review.
Figure 1:
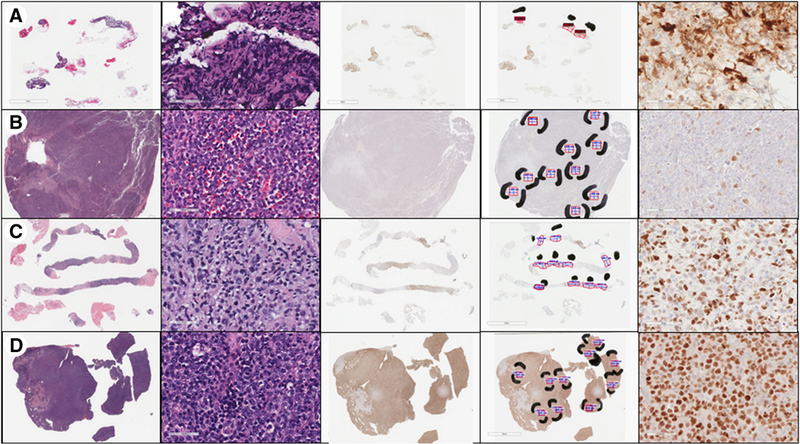
Images from representative cases showing (row A) insufficient material for review, (row B) negative MYC immunohistochemical (IHC) staining, (row C) equivocal MYC IHC staining (percent positivity in 30-50% range), and (row D) positive MYC IHC staining. The first column shows whole-slide images of the hematoxylin and eosin (H&E) stained slide. The second column shows a portion of the H&E slide at 400x magnification, in one of the selected areas of best tissue. The third column demonstrates whole-slide images of the MYC IHC slide. The fourth column shows the Aperio ImageScope annotations on the MYC IHC slide, with black marking pen indicating the areas of best quality tissue selected as the areas to evaluate. The red boxes select tissue to analyze, while the text and ruler icons in blue measure the area selected, with the goal of 1 mm2 in each area. The fifth column shows the MYC IHC slide at 400x magnification, within one of the evaluation areas.
For the retrospective review by digital image analysis, an H&E and MYC immunohistochemical slide for each case were scanned at 40x using an Aperio ScanScope XT whole slide scanner (Leica Biosystems, Buffalo Grove, IL), including on-slide control tissue. These were converted to digital images and stored on a password-protected database using Aperio eSlide Manager (version 12.3, Leica Biosystems). The MYC stained slides were annotated using Aperio ImageScope viewing software (version 12.3, Leica Biosystems). The ruler tool was used to measure 1 mm x 1 mm squares in each area previously marked by dotting pen on the slide, then the free-hand pen tool or rectangle tool were used to annotate areas for analysis. The annotations on each MYC stained slide were analyzed using the Aperio Nuclear algorithm (version 9.2, Leica Biosystems), with no manipulation to the algorithm. Default settings for the nuclear algorithm were used (“Nuclear Algorithm, User’s Guide” Leica Biosystems, MAN-0338, Revision 8; August 5, 2015). All data was saved individually by case, and the numbers of negative, 1+, 2+, and 3+ nuclei were specifically collated. We did not utilize the negative versus positive (1+ through 3+) pixel count for this study.
For the manual retrospective review, a university based hematopathologist reviewer (pathologist #1) graded the percent of cells with positive nuclear staining for each area previously marked by dotting pen on the MYC stained slide. The percentage of nuclei with brightly positive staining was recorded as well as the percentage of nuclei with any staining (that is, dim and bright alike). An average from all marked areas (up to 10 per slide) was calculated to obtain separate percent positive staining values for dim staining and any staining. When the final value for percent positivity fell between 30% and 50% by manual retrospective review, a second university based hematopathologist reviewer (pathologist #2) performed the same grading process. On-slide control tissue and the corresponding H&E stained tissue slide were evaluated as needed. Pathologists #1 and #2 were blinded to the retrospective digital image analysis, which was performed by a third pathologist.
To test the association between MYC gene status and variables of interest, Wilcoxon rank-sum tests and Fisher exact tests were performed, as appropriate. Correlation was evaluated using Spearman correlation coefficients and t-tests. Concordance was evaluated using the kappa statistic(10). Specificities and sensitivities were calculated. All reported p-values are two-sided. Statistical analyses were performed using SAS 9.4 (SAS Institute, Inc., Cary, NC) and R version 3.4.0 (http://www.R-project.org/).
3. Results
3.1. General characteristics
Our cohort consisted of 87 cases: 72 (83%) DLBCL or high-grade B-cell lymphoma (as defined by 2016 WHO classification), 12 (14%) monomorphic PTLD, and 3 (3%) Burkitt lymphoma (Figure 2). Patients ranged in age from 3 to 93 years old at the time of biopsy with an average age of 58. More of the biopsies were from female than male patients (50 female: 37 male). Sampled sites included: lymph node (n=34), soft tissue and bone (26), oropharynx/sinonasal (7), gastrointestinal tract (4), liver (4), brain (4), gonadal (3), and one each from salivary gland, breast, omentum, thyroid, and lung. Thirty-four cases were core biopsies and 53 were excisional biopsies. For the manual retrospective review and retrospective review by digital image analysis, 32 samples had less than the goal of 10 areas evaluated, of these, 19 were core biopsies and 13 were excisional biopsies. For these 32 samples, an average of 5.2 areas were evaluated.
Figure 2:
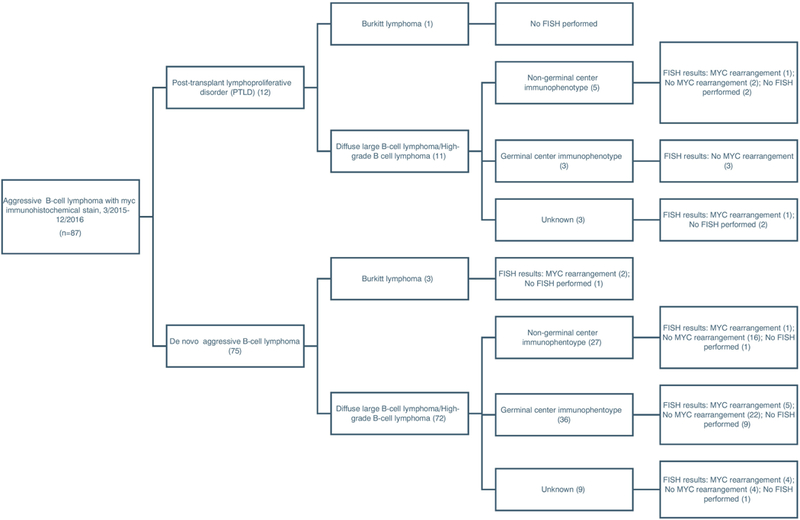
Flow chart outlining the classification of aggressive B-cell lymphomas included in our study.
For each case, the original IHC interpretation was made as part of routine patient care by one of twelve different pathologists - six fellowship-trained hematopathologists practicing at a university hospital and six community-based pathologists (some with hematopathology fellowship training). The majority of cases were originally signed out by the university-based hematopathologists (72%) with the remainder signed out by community-based pathologists. We saw no pattern differences in MYC interpretation between the university hospital and community based pathologists (data not shown).
3.2. Distribution and correlation of percent MYC positivity
Figure 3 illustrates the distribution of percent MYC positivity by retrospective manual review (pathologist #1) and retrospective digital image analysis for any staining and 1+ to 3+, respectively. See Supplemental Material for the distribution of bright staining only by retrospective manual review and 2+ to 3+ by digital image analysis. It can be seen there is a wide spectrum of MYC percent positivity by both methods. There was a strong positive correlation for MYC percent positivity between manual retrospective review by pathologist #1 and retrospective review by digital image analysis with a Spearman correlation coefficient of approximately 0.90 (p<0.01) for all comparisons (manual any staining, manual bright staining versus digital image analysis 1+ to 3+, digital image analysis 2+ to 3+). A representative scatterplot is shown in Figure 4. When we restricted analysis to the 22 cases with a percent MYC positivity between 30% and 50% (as determined by manual retrospective review by pathologist #1), the correlation to retrospective review by digital image analysis was reduced with a Spearman correlation coefficient of 0.438, and there was no correlation between pathologist #1 and pathologist #2 (Spearman correlation coefficient of 0.236, p=0.291).
Figure 3:
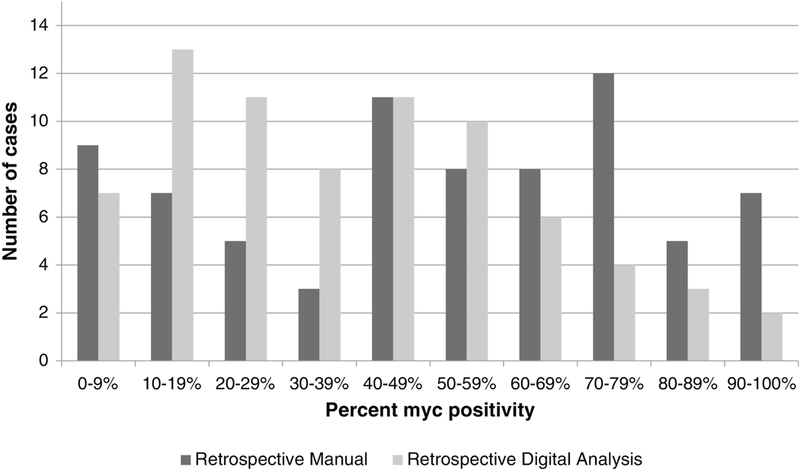
Distribution of percent MYC positivity by retrospective manual review (pathologist #1) and retrospective digital image analysis. Represented in this figure are the percentages using any nuclear staining (retrospective manual) and 1+ to 3+ nuclear positivity (retrospective digital analysis). Both the manual and digital analysis methods show wide variability in percent MYC positivity, including many cases surrounding the suggested cut-off of 40%. The manual method showed a bias toward higher estimation of MYC percent positivity, while the digital analysis method showed a bias toward lower estimation of MYC percent positivity.
Figure 4:
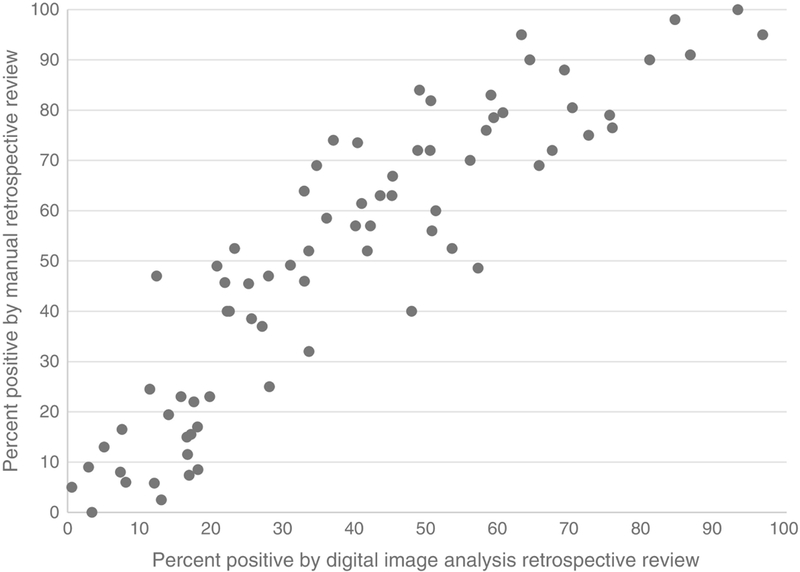
Scatterplot demonstrating correlation between retrospective manual review of MYC immunohistochemistry positivity and digital review by Aperio ImageScope with nuclear algorithm (Spearman correlation coefficient of 0.92).
3.3. Concordance among methods of MYC immunohistochemical staining evaluation
The original sign-out pathologist interpreted the MYC immunohistochemical stain as positive in 41 cases (47%), negative in 37 (43%), and equivocal in 8 (9%), with an interpretation not given in 1 (1%). Using a cut-off of ≥ 40% as positive, 51 of the 75 cases (68%) deemed sufficient material for evaluation by the manual retrospective method were positive for any nuclear staining and 30 of the 75 cases (40%) were positive for bright nuclear staining. Twelve cases were deemed insufficient material to evaluate by the manual retrospective method but were evaluated by retrospective digital image analysis. Using a cut-off of ≥ 40% as positive, forty of the 87 cases (46%) evaluated by retrospective digital image analysis were positive when 1+ through 3+ nuclear positivity was interpreted as positive and 12 of the 87 cases (14%) evaluated by retrospective digital image analysis were positive when 2+ through 3+ nuclear positivity was interpreted as positive.
Concordance values among the three methods of MYC immunohistochemical stain evaluation (comparing interpretation of “positive” or “negative”) are shown in Table 1. There was excellent concordance between the retrospective manual review and the digital image analysis on the core biopsy specimens; there was good concordance between all other comparisons. When evaluation was limited to the subset with staining in the 30-50% range (all specimens), concordance values were 50%, 60%, and 45% respectively for original IHC interpretation to manual retrospective review, original IHC interpretation to retrospective review by digital image analysis, and retrospective manual review to retrospective review by digital image analysis. Corresponding kappa scores were 0.138, 0.130, and 0.137, respectively,, indicating marginal/poor agreement. There was marginal/poor agreement between pathologist #1 and pathologist #2 for the subset of cases staining in the 30-50% range.
Table 1:
Concordance Among Methods of MYC Immunohistochemical Stain Evaluation1
| Concordance | Cohen Kappa Statistic | Agreement2 | |
|---|---|---|---|
| Original IHC interpretation to manual retrospective review (n=69) | 80% | 0.588 | Good |
| Original IHC interpretation to retrospective review by digital image analysis (n=78) | 81% | 0.616 | Good |
| Retrospective manual review to retrospective review by digital image analysis (n=75) | 80% | 0.606 | Good |
| Core Biopsy Only | |||
| Original IHC interpretation to manual retrospective review (n=23) | 87% | 0.738 | Good |
| Original IHC interpretation to retrospective review by digital image analysis (n=30) | 80% | 0.598 | Good |
| Retrospective manual review to retrospective review by digital image analysis (n=24) | 96% | 0.915 | Excellent |
| Excisional Biopsy Only | |||
| Original IHC interpretation to manual retrospective review (n=46) | 76% | 0.513 | Good |
| Original IHC interpretation to retrospective review by digital image analysis (n=48) | 81% | 0.626 | Good |
| Retrospective manual review to retrospective review by digital image analysis (n=51) | 73% | 0.474 | Good |
Concordance evaluated using any nuclear staining by manual retrospective review and 1+ through 3+ nuclear positivity by digital image analysis to determine percent myc positivity, and using a cut-off of ≥ 40% as positive.
As defined by Le (10).
3.4. MYC immunohistochemical stain versus MYC gene status
Sixty-one cases were evaluated by FISH for MYC gene rearrangement on the same tissue sample as the MYC immunohistochemical stain. Fourteen of 61 tested cases (23%) had a MYC rearrangement by FISH analysis. A summary of MYC immunohistochemical staining (as evaluated by the various methods) compared to the presence or absence of a MYC gene rearrangement in excisional biopsy specimens is presented in Table 2. There was a significant association between MYC immunohistochemical staining and MYC gene rearrangement status for all methods when assessing excisional biopsy specimens alone and when assessing all specimens together (core and excisional biopsies, p<0.01). However, when evaluation was restricted to core biopsy specimens only (24 cases evaluated by FISH, 5 with MYC gene rearrangement detected), no significant results of association were found. A significant association was not identified between any of the variables and MYC gene gain/amplification. Using a cut-off of ≥ 40% as positive, Table 3 shows the sensitivity and specificity for the various methods of MYC immunohistochemical staining evaluation in predicting MYC gene rearrangement by FISH analysis. In Table 3, data for core biopsies and excisional specimens are presented together (“All Specimens”) and separately.
Table 2.
MYC immunohistochemical staining versus MYC gene rearrangement status (Excisional biopsy specimens only)
|
MYC
gene rearrangement (N=9)1 |
No MYC gene rearrangement (N=28)1 |
P- value |
|
|---|---|---|---|
| Myc immunostain percent positivity by: | Median (25%, 75%) | Median (25%, 75%) | |
| Digital image analysis (1++−3+) | 65.6 (63.1, 80.9) | 29.5 (14.7, 43.4) | 0.001 |
| Digital image analysis (2++−3+) | 30.7 (19.3, 51.3) | 14.0 (5.3, 23.8) | 0.02 |
| Manual retrospective (any staining) | 90.0(81.9, 91.0) | 48.6 (23.0, 63.9) | 0.0005 |
| Manual retrospective (bright staining) | 67.5 (38.1, 85.0) | 25.0 (9.0, 42.8) | 0.009 |
| Original pathologist’s interpretation of myc staining: | Number (percentage) | Number (percentage) | 0.002 |
| Positive | 9 (100) | 9 (32.1) | |
| Negative | 0 (0.0) | 15 (53.6) | |
| Equivocal | 0 (0.0) | 4 (14.3) |
Overall N may vary by variable.
Table 3:
Sensitivity and specificity of the various methods of MYC immunohistochemical stain interpretation for MYC rearrangement determination
| Original IHC Interpretation | ||||
| Sensitivity | Specificity | |||
| All Specimens (n=54) | 0.93 | 0.65 | ||
| Germinal Center Subtype (n=28) | 0.80 | 0.70 | ||
| Non-germinal center subtype (n=16) | 1.00 | 0.57 | ||
| Core (n=21) | 0.80 | 0.69 | ||
| Excision (n=33) | 1.00 | 0.63 | ||
| Manual Retrospective Review1 | ||||
| Sensitivity | Specificity | |||
| Any | Bright | Any | Bright | |
| All Specimens (n=55) | 0.92 | 0.62 | 0.40 | 0.69 |
| Core (n=19) | 0.75 | 0.50 | 0.53 | 0.67 |
| Excision (n=36) | 1.00 | 0.67 | 0.33 | 0.70 |
| Retrospective Review by Digital Image Analysis2 | ||||
| Sensitivity | Specificity | |||
| Any | Bright | Any | Bright | |
| All Specimens (n=61) | 0.86 | 0.50 | 0.66 | 0.98 |
| Core (n=24) | 0.80 | 0.60 | 0.63 | 1.00 |
| Excision (n=37) | 0.89 | 0.44 | 0.68 | 0.96 |
Note: Of the 61 cases with FISH analysis, 6 were deemed insufficient to evaluate by retrospective manual review and 7 were deemed equivocal by original pathologist’s interpretation, accounting for varying “n”.
For the manual retrospective review, “Any” refers to MYC ihc percent positivity coming from nuclear staining of any intensity (bright or dim) and “Bright” refers to MYC ihc percent positivity coming from nuclear staining of only bright intensity.
For the digital image analysis, “Any” refers to MYC ihc percent positivity coming from 1+ through 3+ nuclear positivity and “Bright” refers to MYC ihc percent positivity coming from 2+ through 3+ positivity.
4. Discussion
Original reports in the literature on interpretation of the MYC immunohistochemical stain reported high concordance rates among pathologists; however, those studies were based on tissue microarrays and/or review among few pathologists at single institutions (2, 11-13). Subsequent studies have shown less concordance among pathologists when whole tissue sections were scored by a diverse group of pathologists (6, 14). Scoring of the MYC immunohistochemical stain can be particularly difficult due to both staining heterogeneity between areas on whole tissue sections and heterogeneity of staining intensity. There is currently no guidance on how to best interpret staining heterogeneity. In our study, we used a systematic approach to retrospective review in an attempt to compensate for staining heterogeneity. In addition, we used digital image analysis as an alternative, and perhaps less biased, method of evaluating MYC immunohistochemical staining(15) (16). The percentage of MYC positivity between our manual retrospective review and retrospective review by digital image analysis showed strong correlation (Spearman correlation coefficients of approximately 0.90 for all comparisons). However, when the data was dichotomized to positive or negative for MYC based on a cut-off of 40%, concordance between the three methods of evaluation (original IHC interpretation, manual retrospective review, and retrospective review by digital image analysis) was only around 80%, with Kappa scores showing good agreement. This concordance is similar to that reported among nine different pathologists in Mahmoud, et al (6). Interestingly, concordance for the core biopsy specimens between the retrospective manual review and retrospective review by digital image analysis in our study was excellent (96%, kappa score 0.915), perhaps due to fewer areas evaluated in these specimens or better fixation of the specimen.
In our series, core biopsy specimens were poor predictors of MYC gene rearrangement. When we evaluated the core biopsy specimens in isolation, no significant results of association were found between MYC immunohistochemical stain and MYC gene rearrangement, and sensitivities using a cut-off of 40% positivity were only 0.50 to 0.80 by the various methods used for evaluation. The sensitivity of excisional biopsy specimens was excellent by the original pathologist’s interpretation and by manual retrospective review, at the expense of specificity (sensitivities of 1.0 and specificities of 0.63 and 0.33, respectively). The drastic decrease in specificity when evaluated by manual retrospective review (performed in a more systematic fashion and not influenced by morphology and results of additional immunohistochemical stains) suggests that, in a clinical setting, our pathologists used other clues to predict a MYC gene rearrangement, such as clinical behavior, Ki-67 proliferation index, and aggressive morphology. Alternatively, this could represent the individual bias of the retrospective pathologist reviewer to overestimate percent positivity in this setting. It has been recommended in the literature that MYC immunohistochemical staining should not be used as the sole method of MYC status evaluation (14). Our study findings strongly support this conclusion for core biopsy specimens. For excisional biopsy specimens, MYC immunohistochemical stain may be a triaging tool if used with great caution. Correlating MYC immunohistochemical stain results with morphologic and immunohistochemical features suggestive of Burkitt lymphoma or high grade B-cell lymphoma is invaluable. We found no association between MYC immunohistochemical stain and MYC gene gain/amplification, although we may not have had enough cases to identify an association.
In our study, digital image analysis had a similar sensitivity for MYC rearrangement to the original pathologist’s interpretation and the retrospective manual interpretation with a similar specificity to the original pathologist’s interpretation when all specimens were evaluated, although it did have a slightly lower sensitivity (0.89) when core biopsy specimens were excluded. The concordance between retrospective review by digital image analysis and the original IHC interpretation was similar to the previously reported concordance among pathologists and the correlation between our manual retrospective review and retrospective review by digital image analysis was strong(6). These findings indicate that digital image analysis performs comparably to manual analysis, and therefore is a viable alternative to manual analysis for interpretation of MYC immunohistochemical staining. Our method used the Aperio Nuclear algorithm without manipulation, a strategy that would need confirmation across other institutions. Advantages to digital image analysis include the possibility of a more standardized approach to interpretation of the immunohistochemical stain, the opportunity for a more sophisticated scoring system (for example, taking into account the relative distribution of nuclei with different staining intensities) and the ability of the interpretation to occur at a central location(7). Disadvantages include the processing power and time required for scanning and analyzing the slides. Our approach to digital image analysis still required manual input to identify viable areas of neoplastic tissue. Applying the nuclear algorithm to the whole slide without manual input would include non-malignant cells (endothelial cells, non-neoplastic lymphocytes, fibroblasts, etc.) in the analysis and introduce artifact from crush, overstaining, tissue folds, and degenerating neoplastic cells.
Beyond its use as a potential triaging tool, expression of MYC protein as determined by immunohistochemical staining is a potential prognostic and/or predictive marker. Double expression of MYC and BCL2 proteins in diffuse large B-cell lymphoma, not otherwise specified (DLBCL, NOS) is associated with inferior survival in most studies (2, 4, 12, 17-21). Most studies, but not all, use the cut-off of 40% as indicative of MYC positivity or “overexpression.” In our cohort, nearly one-third of our cases had a percent MYC positivity surrounding the cut-off of 40% and, for these cases, our concordance values among the various methods used for evaluation was marginal/poor. This suggests there is limited utility of this stain as a prognostic or predictive marker using 40% as a cut-off value without further evaluation guidelines. Tsuyama et al. performed a systematic review of BCL2 immunohistochemistry using a scoring system for staining of 0 to 3+, dependent on proportion of cells staining and intensity of staining, and found that a BCL2 3+ score was a significant prognostic factor (22). A similar scoring system may prove to be valuable for MYC immunohistochemical stain interpretation. Digital image analysis may be a robust method to develop a more sophisticated scoring system, standardize the interpretation, and allow for centralized review. Our study was not designed to evaluate clinical outcome; however, we feel that our study demonstrates comparable performance of digital image analysis to manual review, therefore suggesting that future studies can incorporate digital image analysis. Clear guidelines for evaluation of MYC immunohistochemical staining interpretation is recommended for future studies regarding its prognostic or predictive value.
To summarize, our results suggest that the MYC immunohistochemical stain should not be used as a triaging tool for the MYC gene rearrangement in core biopsy specimens. For excisional biopsy specimens, there is potential use as a triaging tool with the caveat that the sensitivity is not 100% (as per the literature and our experience during validation of the MYC immunohistochemical stain in our laboratory) and the acknowledgement that there is a low specificity. Results of the MYC immunohistochemical stain should always be correlated with morphologic and clinical findings and results of other immunohistochemical stains. If the MYC immunohistochemical stain is to be used as a prognostic or predictive marker, clear guidelines need to be developed delineating the best method of stain interpretation for this purpose, and digital image analysis should be investigated as a potential method of stain interpretation..
Supplementary Material
MYC IHC on core biopsies is a poor predictor of MYC rearrangement.
The MYC IHC stain has limited prognostic utility without further guidelines.
Digital image analysis (DIA) is a viable alternative method of MYC IHC evaluation
DIA may be especially useful in future prognostic/predictive studies.
Acknowledgments
Research reported in this publication was supported by NIH grant P30 CA77598 utilizing the Biostatistics and Bioinformatics Core shared resource of the Masonic Cancer Center, University of Minnesota and by the National Center for Advancing Translational Sciences of the National Institutes of Health Award Number UL1TR000114. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Disclosures/Conflicts of Interest: The authors have no conflicts of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Gascoyne RD, Campo E, Jaffe ES, Chan WC, Chan JKC, Rosenwald A, Stein H, Swerdlow SH. Diffuse large B-cell lymphoma, NOS In: Swerdlow SH, Campo E, Harris NL, Jaffe ES, Pileri SA, Stein H, Thiele J, Arber DA, Hasserjian RP, Le Beau MM, Orazi A, Siebert R, eds. World Health Organization Classification of Tumours of Haematopoietic and Lymphoid Tissue. Lyon: IARC, 2017. [Google Scholar]
- 2.Johnson NA, Slack GW, Savage KJ, Connors JM, Ben-Neriah S, Rogic S, Scott DW, Tan KL, Steidl C, Sehn LH, Chan WC, Iqbal J, Meyer PN, Lenz G, Wright G, Rimsza LM, Valentino C, Brunhoeber P, Grogan TM, Braziel RM, Cook JR, Tubbs RR, Weisenburger DD, Campo E, Rosenwald A, Ott G, Delabie J, Holcroft C, Jaffe ES, Staudt LM, Gascoyne RD. Concurrent expression of MYC and BCL2 in diffuse large B-cell lymphoma treated with rituximab plus cyclophosphamide, doxorubicin, vincristine, and prednisone. J Clin Oncol 2012; 30, 3452–3459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Petrich AM, Nabhan C, Smith SM. MYC-associated and double-hit lymphomas: a review of pathobiology, prognosis, and therapeutic approaches. Cancer 2014; 120, 3884–3895. [DOI] [PubMed] [Google Scholar]
- 4.Horn H, Ziepert M, Becher C, Barth TF, Bernd HW, Feller AC, Klapper W, Hummel M, Stein H, Hansmann ML, Schmelter C, Moller P, Cogliatti S, Pfreundschuh M, Schmitz N, Trumper L, Siebert R, Loeffler M, Rosenwald A, Ott G, German High-Grade Non-Hodgkin Lymphoma Study G. MYC status in concert with BCL2 and BCL6 expression predicts outcome in diffuse large B-cell lymphoma. Blood 2013; 121, 2253–2263. [DOI] [PubMed] [Google Scholar]
- 5.Fitzgibbons PL, Bradley LA, Fatheree LA, Alsabeh R, Fulton RS, Goldsmith JD, Haas TS, Karabakhtsian RG, Loykasek PA, Marolt MJ, Shen SS, Smith AT, Swanson PE, College of American Pathologists P, Laboratory Quality C. Principles of analytic validation of immunohistochemical assays: Guideline from the College of American Pathologists Pathology and Laboratory Quality Center. Arch Pathol Lab Med 2014; 138, 1432–1443. [DOI] [PubMed] [Google Scholar]
- 6.Mahmoud AZ, George TI, Czuchlewski DR, Zhang QY, Wilson CS, Sever CE, Bakhirev AG, Zhang D, Steidler NL, Reichard KK, Kang H, Foucar K, Vasef MA. Scoring of MYC protein expression in diffuse large B-cell lymphomas: concordance rate among hematopathologists. Mod Pathol 2015; 28, 545–551. [DOI] [PubMed] [Google Scholar]
- 7.Ghaznavi F, Evans A, Madabhushi A, Feldman M. Digital imaging in pathology: whole-slide imaging and beyond. Annu Rev Pathol 2013; 8, 331–359. [DOI] [PubMed] [Google Scholar]
- 8.Marcuzzo T, Giudici F, Ober E, Rizzardi C, Bottin C, Zanconati F. Her2 immunohistochemical evaluation by traditional microscopy and by digital analysis, and the consequences for FISH testing. Pathol Res Pract 2016; 212, 911–918. [DOI] [PubMed] [Google Scholar]
- 9.Rizzardi AE, Zhang X, Vogel RI, Kolb S, Geybels MS, Leung YK, Henriksen JC, Ho SM, Kwak J, Stanford JL, Schmechel SC. Quantitative comparison and reproducibility of pathologist scoring and digital image analysis of estrogen receptor beta2 immunohistochemistry in prostate cancer. Diagn Pathol 2016; 11, 63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Le CT. Applied categorical data analysis and translational research. Hoboken, NJ: Wiley, 2010. [Google Scholar]
- 11.Kluk MJ, Chapuy B, Sinha P, Roy A, Dal Cin P, Neuberg DS, Monti S, Pinkus GS, Shipp MA, Rodig SJ. Immunohistochemical detection of MYC-driven diffuse large B-cell lymphomas. PLoS One 2012; 7, e33813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Perry AM, Alvarado-Bernal Y, Laurini JA, Smith LM, Slack GW, Tan KL, Sehn LH, Fu K, Aoun P, Greiner TC, Chan WC, Bierman PJ, Bociek RG, Armitage JO, Vose JM, Gascoyne RD, Weisenburger DD. MYC and BCL2 protein expression predicts survival in patients with diffuse large B-cell lymphoma treated with rituximab. Br J Haematol 2014; 165, 382–391. [DOI] [PubMed] [Google Scholar]
- 13.Green TM, Nielsen O, de Stricker K, Xu-Monette ZY, Young KH, Moller MB. High levels of nuclear MYC protein predict the presence of MYC rearrangement in diffuse large B-cell lymphoma. Am J Surg Pathol 2012; 36, 612–619. [DOI] [PubMed] [Google Scholar]
- 14.Kluk MJ, Ho C, Yu H, Chen BJ, Neuberg DS, Dal Cin P, Woda BA, Pinkus GS, Rodig SJ. MYC Immunohistochemistry to Identify MYC-Driven B-Cell Lymphomas in Clinical Practice. Am J Clin Pathol 2016; 145, 166–179. [DOI] [PubMed] [Google Scholar]
- 15.Sander B, de Jong D, Rosenwald A, Xie W, Balague O, Calaminici M, Carreras J, Gaulard P, Gribben J, Hagenbeek A, Kersten MJ, Molina TJ, Lee A, Montes-Moreno S, Ott G, Raemaekers J, Salles G, Sehn L, Thorns C, Wahlin BE, Gascoyne RD, Weller E. The reliability of immunohistochemical analysis of the tumor microenvironment in follicular lymphoma: a validation study from the Lunenburg Lymphoma Biomarker Consortium. Haematologica 2014; 99, 715–725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Aeffner F, Wilson K, Martin NT, Black JC, Hendriks CLL, Bolon B, Rudmann DG, Gianani R, Koegler SR, Krueger J, Young GD. The Gold Standard Paradox in Digital Image Analysis: Manual Versus Automated Scoring as Ground Truth. Arch Pathol Lab Med 2017; 141, 1267–1275. [DOI] [PubMed] [Google Scholar]
- 17.Rosenthal A, Younes A. High grade B-cell lymphoma with rearrangements of MYC and BCL2 and/or BCL6: Double hit and triple hit lymphomas and double expressing lymphoma. Blood Rev 2017; 31, 37–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hu S, Xu-Monette ZY, Tzankov A, Green T, Wu L, Balasubramanyam A, Liu WM, Visco C, Li Y, Miranda RN, Montes-Moreno S, Dybkaer K, Chiu A, Orazi A, Zu Y, Bhagat G, Richards KL, Hsi ED, Choi WW, Zhao X, van Krieken JH, Huang Q, Huh J, Ai W, Ponzoni M, Ferreri AJ, Zhou F, Slack GW, Gascoyne RD, Tu M, Variakojis D, Chen W, Go RS, Piris MA, Moller MB, Medeiros LJ, Young KH. MYC/BCL2 protein coexpression contributes to the inferior survival of activated B-cell subtype of diffuse large B-cell lymphoma and demonstrates high-risk gene expression signatures: a report from The International DLBCL Rituximab-CHOP Consortium Program. Blood 2013; 121, 4021–4031; quiz 4250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Green TM, Young KH, Visco C, Xu-Monette ZY, Orazi A, Go RS, Nielsen O, Gadeberg OV, Mourits- Andersen T, Frederiksen M, Pedersen LM, Moller MB. Immunohistochemical double-hit score is a strong predictor of outcome in patients with diffuse large B-cell lymphoma treated with rituximab plus cyclophosphamide, doxorubicin, vincristine, and prednisone. J Clin Oncol 2012; 30, 3460–3467. [DOI] [PubMed] [Google Scholar]
- 20.Valera A, Lopez-Guillermo A, Cardesa-Salzmann T, Climent F, Gonzalez-Barca E, Mercadal S, Espinosa I, Novelli S, Briones J, Mate JL, Salamero O, Sancho JM, Arenillas L, Serrano S, Erill N, Martinez D, Castillo P, Rovira J, Martinez A, Campo E, Colomo L, Grup per l’Estudi dels Limfomes de Catalunya i B. MYC protein expression and genetic alterations have prognostic impact in patients with diffuse large B- cell lymphoma treated with immunochemotherapy. Haematologica 2013; 98, 1554–1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Scott DW, Mottok A, Ennishi D, Wright GW, Farinha P, Ben-Neriah S, Kridel R, Barry GS, Hother C, Abrisqueta P, Boyle M, Meissner B, Telenius A, Savage KJ, Sehn LH, Slack GW, Steidl C, Staudt LM, Connors JM, Rimsza LM, Gascoyne RD. Prognostic Significance of Diffuse Large B-Cell Lymphoma Cell of Origin Determined by Digital Gene Expression in Formalin-Fixed Paraffin-Embedded Tissue Biopsies. J Clin Oncol 2015; 33, 2848–2856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tsuyama N, Sakata S, Baba S, Mishima Y, Nishimura N, Ueda K, Yokoyama M, Terui Y, Hatake K, Kitagawa M, Ishizuka N, Tomita N, Takeuchi K. BCL2 expression in DLBCL: reappraisal of immunohistochemistry with new criteria for therapeutic biomarker evaluation. Blood 2017; 130, 489–500. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


