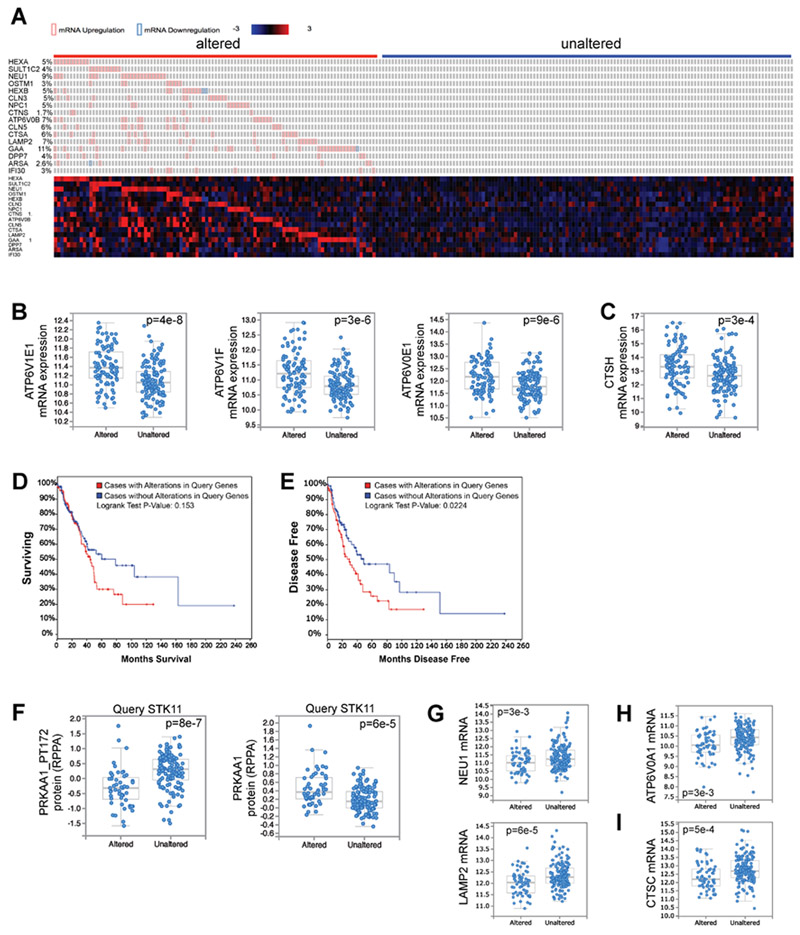
Figure 7. Elevated lysosomal gene expression correlates with accelerated disease recurrence in human lung adenocarcinoma patients.
(A) The 230 complete tumors within the Lung Adenocarcinoma TCGA dataset (TCGA, Provisional, 522 samples) were queried for alterations in mRNA levels of any of the 16 AMPK-dependent lysosomal lung cancer gene signature genes defined in Figure 3I. The RNA-seq z-score cut-off was set at +/− 2. Patient samples were divided into two groups; those with modulation of mRNA levels of lysosomal gene signature genes, termed “altered,” or those without major mRNA alterations in any of these genes, termed “unaltered.” Heatmap illustrates mRNA expression of each gene across all patient samples.
(B) All 9 V-ATPase genes that were not part of the lysosomal gene signature genes but are significantly misregulated (q-value < 0.05) between the “altered” and “unaltered” tumors are upregulated in the “altered” gene set. 3 examples are shown.
(C) Two cathepsin genes that were not part of the lysosomal gene signature genes are significantly upregulated (q-value < 0.05) in the “altered” gene set. One example is shown.
(D) Overall patient survival after stratification of the data according to alterations in the lysosomal gene signature (A).
(E) Disease free survival after stratification of the data according to alterations in the lysosomal gene signature (A).
(F) The Lung Adenocarcinoma TCGA dataset was queried according to mutation status in STK11, segregating the data into tumors with mutations in STK11, “Altered,” and those without mutations in STK11, “Unaltered”. Plot of AMPK phosphorylation at T172 (PRKAA1_PT172) within the associated proteomics data (left). Total PRKAA1 protein levels (right).
(G) mRNA levels of lysosomal gene signature genes.
(H) mRNA levels of a V-ATPase gene.
(I) mRNA levels of a cathepsin gene.