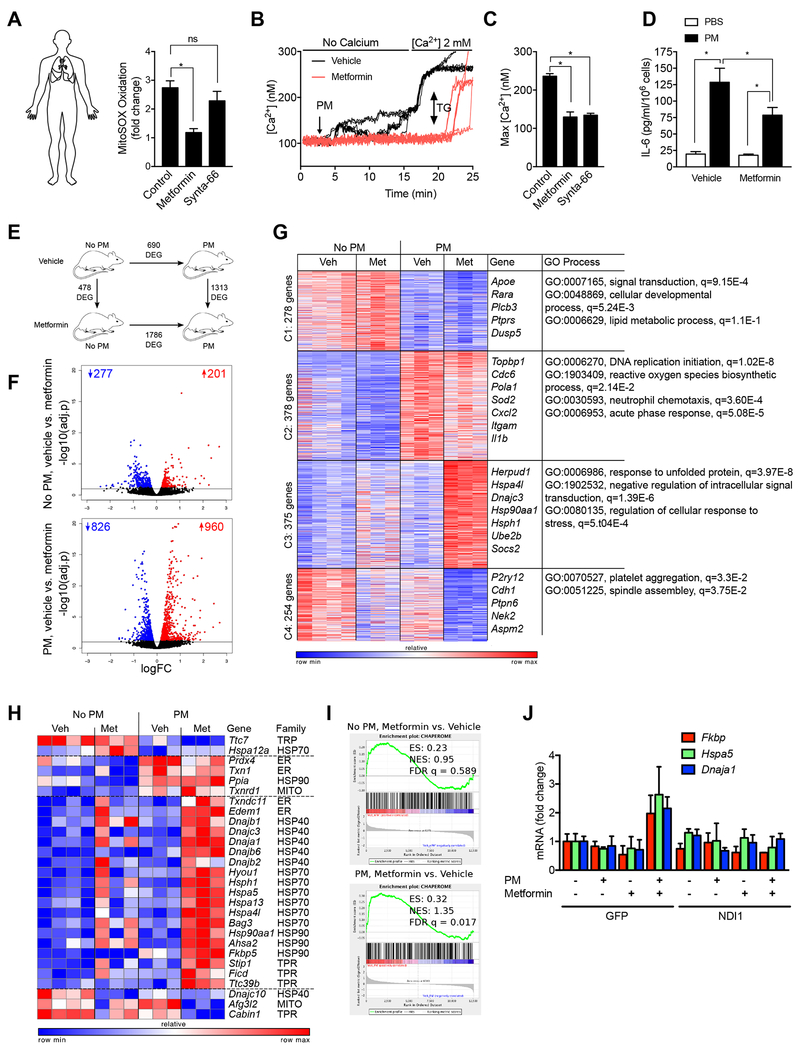
Figure 7. Metformin prevents PM-induced IL-6 release from primary human alveolar macrophages and modifies transcriptional response to PM in murine alveolar macrophages.
(A) Flow sorted alveolar macrophages were isolated from biopsies from donor lungs obtained at the time of lung transplantation and allowed to adhere to glass coverslips for 4-8 hours. The cells were loaded with MitoSOX (5 μM) and then pretreated with saline, metformin (1 mM) or Synta-66 (10 μM) prior to treatment with PM (10 μg/cm2) for measurement of mitochondrial ROS generation. (B,C) Alveolar macrophages were allowed to adhere to glass coverslips prior to loading with Fura-2 (2 μM) and then treated with metformin (1 mM) or saline 1 hour before treatment with PM and intracellular calcium levels were measured in calcium free followed by calcium replete (1 mM) media. Thapsigargin (TG, 25 nM) was added at the end of the experiment. (B) Representative time-series plots and (C) composite data from four individuals is shown. (D) Primary human alveolar macrophages were treated with metformin (1 mM) 1 hour before treatment with PM (10 μg/cm2) and IL-6 levels in the media were measured 4 hours later. (E-G) Mice were treated with metformin in the drinking water for 24 hours before and after instilling PM (10 μg /mouse) intratracheally and alveolar macrophages flow-sorted from whole lung homogenates 24 hours later were subjected to transcriptional profiling via RNA-seq. (E) Schematic of the experimental design. Numbers indicate differentially expressed genes (FDR p < 0.05). (F) Volcano plots demonstrating up- and down-regulated genes in mice treated with metformin before and after exposure to PM, numbers indicate up- and down-regulated genes (FDR p < 0.05). (G) k-means clustering identifies metformin-specific clusters of genes. Differentially expressed genes (1285 genes, identified using ANOVA-like test implemented in edgeR package, FDR p < 0.001) in alveolar macrophages exposed to PM in the presence and absence of metformin pretreatment in vivo were subjected to k-means clustering (number of genes per cluster is shown on the left). Representative gene names and GO processes for each cluster are shown on the right side. (H,I) Chaperones are enriched in cluster 3. Gene Set Enrichment Analysis shows enrichment for chaperone genes after treatment with metformin that is more significant in alveolar macrophages from PM exposed mice. (J) MHS cells stably transfected with a lentivirus encoding NDI1 or a control lentivirus (GFP) were treated with PM and the levels of mRNA encoding the indicated genes were measured 24 hours later using RT-qPCR (n=3, * P<0.05). See also Figure S6.