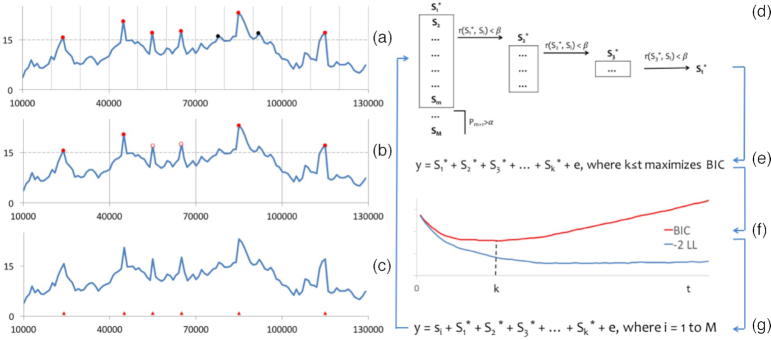
Figure 1:
Limitation of the bin approach and proposed solution. QTNs are rarely distributed evenly throughout the genome, as required by the bin approach used in FarmCPU. The most significant marker from each bin, indicated by the filled red and black circles in (a) and (b), is selected as a pseudo QTN if it passes a threshold (dash lines across vertical axes). A pseudo QTN could be false (filled black circle) if the bins (Separated by the vertical lines) are too small (a) or it could be true but not selected (open circle) if the bins are too big (b), as illustrated by comparing the true QTNs (red triangles) positioned along the horizontal axis in (c). Our alternative method is to sort the M single-nucleotide polymorphisms (SNPs) first and filter them out if their P values are larger than a threshold (α). Among the m SNPs kept, additional SNPs are removed if their correlation (r) with the first SNP (S1*) is larger than a threshold (β). This process is repeated to select S2*, S3*,…, until the last SNP St* is selected (d). As the t remaining SNPs are sorted, we fit the first k of them in a FEM (e) and examine the corresponding twice negative log likelihood (-2LL) and BIC (f). As more SNPs are fitted, -2LL continually improves (blue line), while BIC reverses (red line) because BIC applies a penalty with increasing numbers of SNPs. The set of k SNPs that give the best BIC are used as pseudo QTNs and fitted as covariates in another FEM to test all SNPs, one (si) at a time, as described by the conceptual model (g). This process (d-g) is iterated until the pseudo QTNs remain the same. We named this alternative solution the Bayesian-information and linkage-disequilibrium iteratively nested keyway (BLINK) method.