Abstract
Early in gestation, wounds in fetal skin heal by regeneration, in which microRNAs play key roles. Seq-915_x4024 is a novel microRNA candidate confirmed by deep sequencing and mirTools 2.0. It is highly expressed in fetal keratinocytes during early gestation. Using an in vitro wound-healing assay, Transwell cell migration assay, and MTS proliferation assay, we demonstrated that keratinocytes overexpressing seq-915_x4024 exhibited higher proliferative activity and the ability to promote fibroblast migration and fibroblast proliferation. These characteristics of keratinocytes are the same biological behaviors as those of fetal keratinocytes, which contribute to skin regeneration. In addition, seq-915_x4024 suppressed the expression of the pro-inflammatory markers TNF-α, IL-6, and IL-8 and the pro-inflammatory chemokines CXCL1 and CXCL5. We also demonstrated that seq-915_x4024 regulates TGF-β isoforms and the extracellular matrix. Moreover, using an in vivo wound-healing model, we demonstrated that overexpression of seq-915_x4024 in keratinocytes suppresses inflammatory cell infiltration and scar formation. Using bioinformatics analyses, luciferase reporter assays, and western blotting, we further demonstrated that Sar1A, Smad2, TNF-α, and IL-8 are direct targets of seq-915_x4024. Furthermore, the expression of phosphorylated Smad2 and Smad3 was reduced by seq-915_x4024. Seq-915_x4024 could be used as an anti-fibrotic factor for the treatment of wound healing.
Keywords: microRNA, skin regeneration, keratinocytes, wound healing, scar formation
Introduction
The healing of cutaneous wounds is a highly orchestrated, multistep process that is composed of a complex series of molecular events that prevent infection, repair the damaged tissue, and restore its function.1, 2, 3 Wound healing includes three phases: inflammation, re-epithelialization, and remodeling.4, 5 During the remodeling stage, wounds in adult cutaneous tissues are usually filled with hyperproliferative granulation tissues, which gradually evolve into scar tissues (fibrosis).6
To restore skin tissue, regeneration that leads to scarless healing is the ideal method. Before the third trimester of gestation (early to mid-gestation), wounds in fetal mammalian skin heal rapidly, without scarring or inflammation.7, 8, 9 Then, late-gestational fetal skin changes its response to injury from regeneration to the adult response of fibrosis. This transition occurs in human skin after approximately week 28 of gestation.7 Investigating the mechanism of the transition is of paramount importance to accelerate skin wound healing by changing the response to injury from fibrosis repair to regenerative repair.10
Epidermal keratinocytes (KCs) play important roles in the process of skin tissue regeneration. Early in gestation, fetal KCs exhibit a higher proliferation rate. In addition, fetal KCs enhance fibroblast (FB) migration and FB proliferation and contribute to the extracellular matrix (ECM), which is related to scarless wound healing.11, 12 MicroRNAs (miRNAs), which are a large family of highly conserved, small, noncoding RNAs, play important regulatory roles over a vast number of protein-coding genes.13 Previously, we identified a novel miRNA candidate, seq-915_x4024, which was highly expressed in mid-gestational fetal KCs, and we predicted that seq-915_x4024 may target the transforming growth factor (TGF)-β1-Smad signaling pathway, which plays important roles in skin regeneration.10, 14
In this study, we investigated the roles that seq-915_x4024 plays in wound healing, using the HaCaT cell line (an immortalized human KC cell line). After being transfected with seq-915_x4024 mimics, HaCaT cells exhibited higher proliferative activity and the ability to promote FB migration and FB proliferation. These characteristics of KCs were the same as the biological behaviors of fetal KCs at mid-gestation. In addition, seq-915_x4024 suppressed the pro-inflammatory cytokines tumor necrosis factor-α (TNF-α), interleukin-6 (IL-6), and IL-8 and the pro-inflammatory chemokines CXCL1 and CXCL5. We also demonstrated that seq-915_x4024 regulates the expression of TGF-β isoforms and ECM, including collagen and hyaluronan. Moreover, using an in vivo wound-healing model, we demonstrated that overexpression of seq-915_x4024 in KCs suppresses inflammatory cell infiltration and scar formation. The in vivo expression of collagen type I (Col I), collagen type III (Col III), phosphorylated Smad2 (p-Smad2), and phosphorylated Smad3 (p-Smad3) was suppressed by seq-915_x4024. Furthermore, we used bioinformatics analyses, luciferase reporter assays and western blotting to demonstrate that Sar1A, Smad2, TNF-α, and IL-8 are targets of seq-915_x4024. Overexpression of seq-915_x4024 in KCs suppresses the inflammatory response and scar formation by targeting the TGF-β1-Smad signaling pathway and the inflammatory factors TNF-α and IL-8.
Results
Seq-915_x4024 Regulates Adult KC Biological Behavior
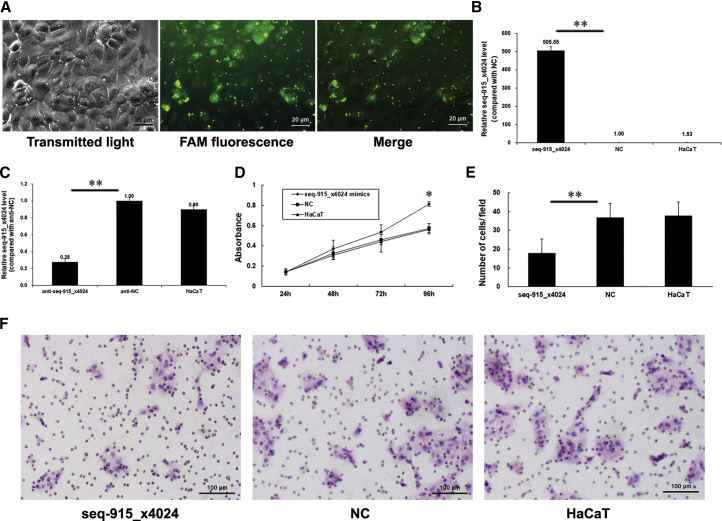
To determine the effect of seq-915_x4024 on regulating the biological behavior of KCs, we transfected an HaCaT cell line with seq-915_x4024 mimics or anti-seq-915_x4024 and their respective negative controls (NCs). Transfection efficiency is shown in Figures 1A–1C. The percentage of FAM-positive HaCaT cells was 94.96% ± 3.76%. Furthermore, we estimated the effects of seq-915_x4024 on the proliferation and migration abilities of HaCaT using the MTS proliferation assay and Transwell cell migration assay. The data demonstrated that seq-915_x4024 exhibits a significant promotional effect on the growth of HaCaT cells (Figure 1D). The results of the Transwell cell migration assay showed that overexpression of seq-915_x4024 obviously inhibits the migration of HaCaT cells. Significantly fewer seq-915_x4024-transfected HaCaT cells (18 ± 7, p < 0.05) passed through the membrane than NC-transfected HaCaT cells (37 ± 7) or parental HaCaT cells (38 ± 7) (Figures 1E and 1F).
Figure 1.
Seq-915_x4024 Regulates Adult KC Biological Behavior
(A) The HaCaT cell line was transfected with 5′-FAM-labeled seq-915_x4024 mimics. The transmitted light image, FAM fluorescence image, and merged image of cells are shown 24 h after transfection. (B) HaCaT cells were transfected with seq-915_x4024 mimics or NCs. Transfection efficiency was detected by real-time qRT-PCR. After cells were transfected with seq-915_x4024 mimics, the expression of seq-915_x4024 increased 505 ± 21-fold. (C) HaCaT cells were transfected with anti-seq-915_x4024 or anti-NC. Transfection efficiency was detected by real-time qRT-PCR. After cells were transfected with anti-seq-915_x4024, the expression of seq-915_x4024 decreased to 28% ± 4%. (D) Seq-915_x4024 promoting HaCaT proliferation was detected by MTS proliferation assay. (E) The numbers of transfected and parental HaCaT cells passing through the Transwell membrane. The number of cells was counted in 16 independent symmetrical visual fields under the microscope from 3 independent experiments (original magnification, ×200). (F) Representative photomicrographs of Transwell results for HaCaT cells were taken under ×100 original magnification. The results represent the means of the values. Bars indicate SD. *p < 0.05 and **p < 0.01, statistical significance between groups.
KCs with Overexpressed Seq-915_x4024 Promote FB Migration and FB Proliferation
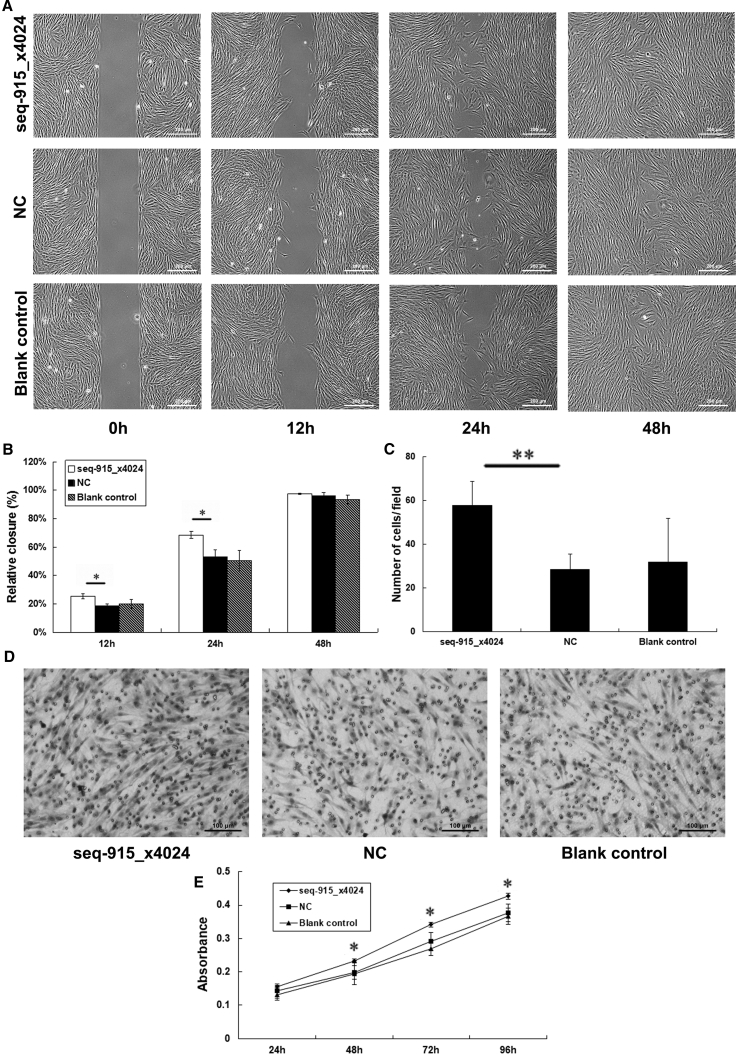
To investigate the effect of seq-915_x4024 in KCs on FB migration, an in vitro wound-healing assay was used. The data demonstrated that, after being cocultured with seq-915_x4024-transfected HaCaT cells, FBs showed a significant increase in cell migration ability (Figures 2A and 2B). In addition, HaCaT cells transfected with anti-seq-915_x4024 significantly inhibited the migration ability of FBs (Figures S1A and S1B).
Figure 2.
KCs with Overexpressed Seq-915_x4024 Promote FB Migration and Proliferation
FBs used for the assays were cocultured with parental or transfected HaCaT cells for 96 h. (A) Representative photomicrographs of FB migration into the scratch wound at 0, 12, 24, and 48 h. (B) Rate of FB movement at 12, 24, and 48 h in the in vitro wound-healing assay. FB migration into the wound area was significantly accelerated in cells cocultured with seq-915_x4024-transfected HaCaT cells compared with the NC group 12 and 24 h after removing the inserts. (C) The number of FBs passing through the Transwell membrane. The number of cells was counted in 16 independent symmetrical visual fields from 3 independent experiments (original magnification, ×200). (D) Representative photomicrographs of Transwell results for FBs were taken under ×100 original magnification. (E) KCs overexpressing seq-915_x4024 promoted FB proliferation as detected by the MTS proliferation assay. The results represent the means of the values. Bars indicate SD. *p < 0.05 and **p < 0.01, statistical significance between groups.
We used a Transwell cell migration assay to further confirm that the overexpression of seq-915_x4024 in KCs promotes FB migration. The data showed that significantly more FBs cocultured with seq-915_x4024-transfected HaCaT cells (58 ± 11, p < 0.01) passed through the membrane than FBs cocultured with NC-transfected HaCaT cells (29 ± 7) and parental HaCaT cells (32 ± 20; Figures 2C and 2D). In contrast, significantly fewer FBs cocultured with anti-seq-915_x4024-transfected HaCaT cells (16 ± 4, p < 0.01) passed through the membrane than FBs cocultured with NC-transfected HaCaT cells (28 ± 5) and parental HaCaT cells (31 ± 7; Figures S1C and S1D).An MTS proliferation assay was used to detect the effect of seq-915_x4024 in KCs on FB proliferation and demonstrated that FBs cocultured with HaCaT cells showed a positive correlation between the expression of seq-915_x4024 in KCs and FB proliferation ability (Figures 2E and S1E).
Seq-915_x4024 Regulates ECM and TGF-β Isoforms
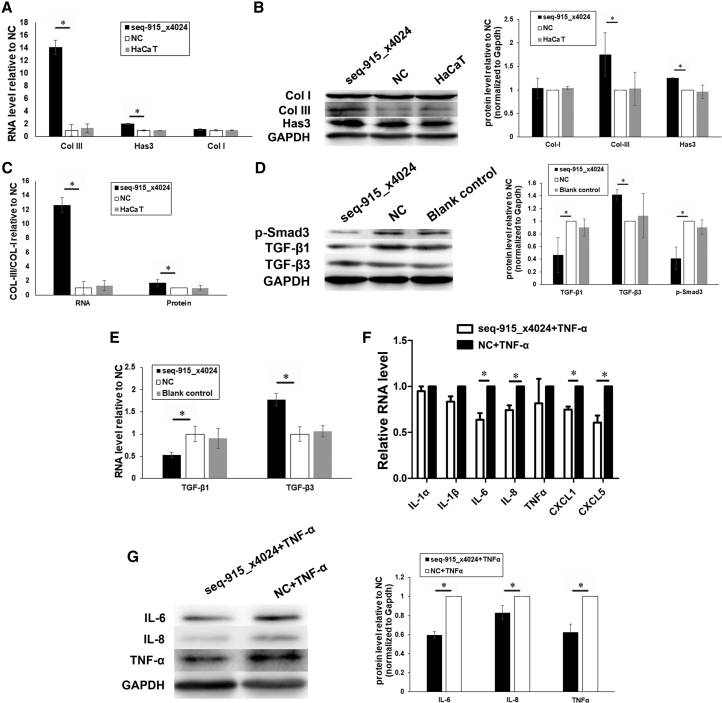
We detected the mRNA and protein levels of the ECM and TGF-β isoforms by real-time qRT-PCR and western blotting. After being transfected with seq-915_x4024 mimics, HaCaT cells expressed more Has3 and Col III compared with cells transfected with NCs (Figures 3A and 3B). In addition, an increased ratio (2.45 ± 0.57 of mRNA and 1.79 ± 0.12 of protein) of type III versus type I collagen was found in seq-915_x4024-transfected HaCaT cells (Figure 3C). FBs cocultured with seq-915_x4024-transfected HaCaT cells expressed less TGF-β1 and p-Smad3 and more TGF-β3 than cells cocultured with NC-transfected or parental HaCaT cells (Figures 3D and 3E).
Figure 3.
Seq-915_x4024 Regulates the ECM, TGF-β Isoforms, and Inflammatory Factors
(A) Real-time qRT-PCR demonstrated that the RNA levels of Col III and Has3 are significantly higher in seq-915_x4024-transfected HaCaT cells than in NC-transfected HaCaT cells. The expression of Col I showed no significant difference. (B) Western blotting confirmed that the expression levels of Col III and Has3 proteins were significantly higher in seq-915_x4024-transfected HaCaT cells compared with those in NC-transfected HaCaT cells. The expression of Col I showed no significant difference. (C) Real-time qRT-PCR and western blotting demonstrated that the ratio of type III versus type I collagen (2.45 ± 0.57 mRNA and 1.79 ± 0.12 protein) was significantly higher in seq-915_x4024-transfected HaCaT cells compared with that in NC-transfected HaCaT cells. (D) Western blotting demonstrated that FBs cocultured with seq-915_x4024-transfected KCs expressed less TGF-β1 and p-Smad3 and more TGF-β3 compared with FBs cocultured with NC-transfected KCs. (E) Real-time qRT-PCR demonstrated that FBs cocultured with seq-915_x4024-transfected KCs expressed less TGF-β1 and more TGF-β3 compared with FBs cocultured with NC-transfected KCs. (F) Real-time qRT-PCR was used to detect the RNA levels of IL-1α, IL-1β, IL-6, IL-8, TNF-α, CXCL1, and CXCL5. The RNA levels of IL-6, IL-8, CXCL1, and CXCL5 were significantly lower in seq-915_x4024-transfected HaCaT cells than in NC-transfected HaCaT cells under inflammatory stimulation. (G) Western blotting showed that under inflammatory stimulation, the expression levels of IL-6, IL-8, and TNF-α in seq-915_x4024-transfected HaCaT cells were significantly lower than those in NC-transfected HaCaT cells. The results represent the means of the values. Bars indicate SD. *p < 0.05, statistical significance between groups.
Seq-915_x4024 Suppresses the Inflammatory Response in KCs
The addition of TNF-α to HaCaT cells was used to reflect inflammatory conditions. HaCaT cells were transfected with seq-915_x4024 mimics or NCs, and then inflammation was induced using TNF-α (20 ng/mL) for 24 h. The expression levels of IL-6, IL-8, CXCL1, and CXCL5 were significantly lower in seq-915_x4024-transfected HaCaT cells compared with levels in NC-transfected HaCaT cells, as detected by real-time qRT-PCR (Figure 3F). Western blot analysis showed that the expression of IL-6, IL-8, and TNF-α was significantly suppressed in seq-915_x4024-transfected HaCaT cells compared with those of NC-transfected HaCaT cells (Figure 3G).
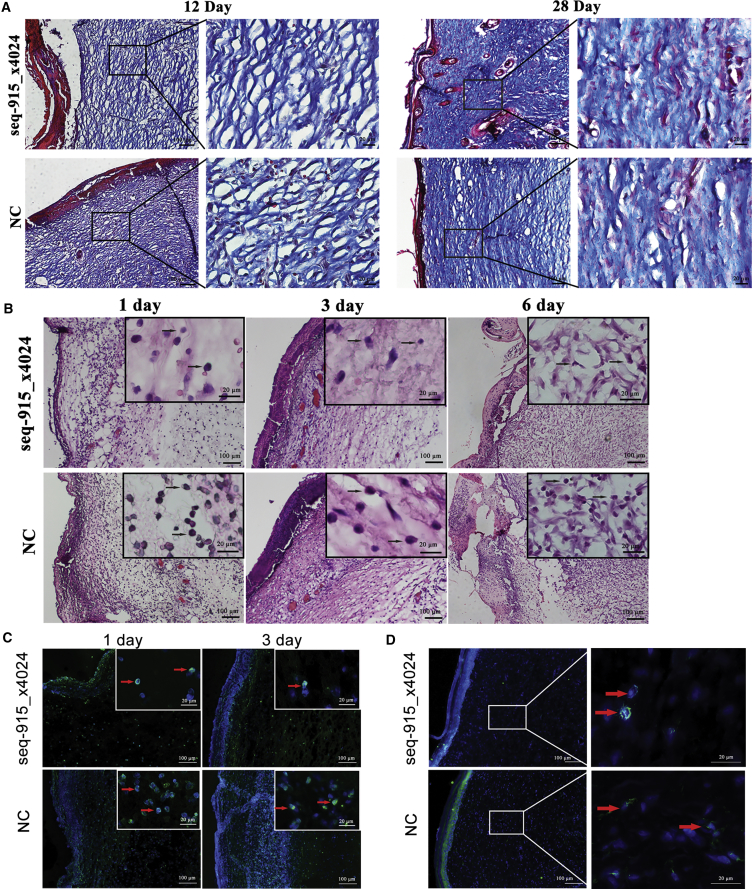
Seq-915_x4024 Suppresses Inflammatory Cell Infiltration and Scar Formation In Vivo
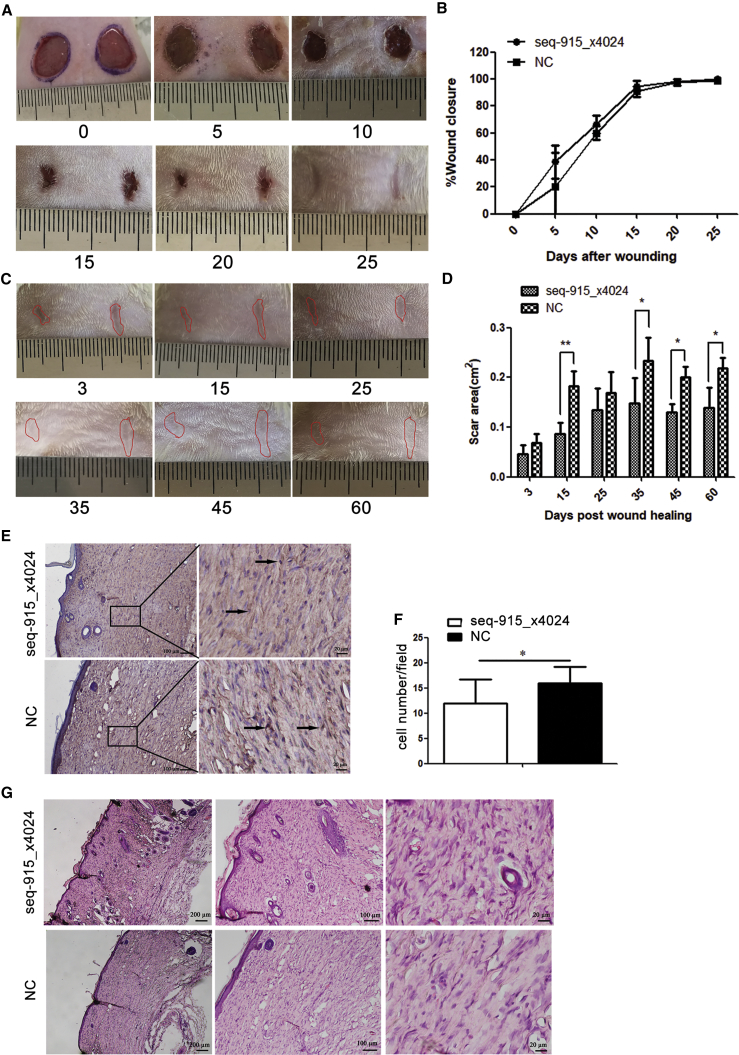
After transplanting the KC cell sheets onto the surface of wounds, we found that overexpression of seq-915_x4024 in KC cell sheets had no significant effect on accelerating wound closure (Figures 4A and 4B). However, on days 15, 35, 45, and 60 after healing, the areas of scar tissue in the seq-915_x4024 group were significantly smaller than those of the NC group. On day 60 after full healing, the scars in the seq-915_x4024 group were 37% ± 12% smaller than those of the control group (Figures 4C and 4D). On day 28 after transplantation, there were fewer α-SMA (α-smooth muscle actin; marker of myofibroblasts)-positive cells, more-organized tissues (skin appendages and new vessels), and uniformly arranged collagen bundles in the deep dermis of the seq-915_x4024 group, than in those of the NC group (Figures 4E–4G and 5A). The wounds on day 12 detected by Masson’s Trichrome staining demonstrated that the closure of open wounds in seq-915_x4024 group rats occurred with deposition of more uniformly organized collagen fiber bundles (Figure 5A). In addition, on days 1, 3, and 6 after transplantation, the inflammatory cell infiltration at the wound sites of the seq-915_x4024 group was significantly lower than that of the NC group (Figures 5B and 5C). At the wound sites on day 28 after transplantation, there were significantly fewer macrophages in the seq-915_x4024 group than in the NC group (Figure 5D).
Figure 4.
KCs with Overexpressed Seq-915_x4024 Suppress Scar Formation In Vivo
KC cell sheets were made of 5 × 106 HaCaT cells seeded into Nunc UpCell (Thermo Fisher) dishes and used in vivo for investigating the effect of seq-915_x4024 on cutaneous regeneration, using a wound-healing model. (A) Photographs of representative wounds in rats on days 0, 5, 10, 15, 20, and 25 after transplantation. (B) Time course of wound closure for each experimental and control group. (C) Representative scars on days 3, 15, 25, 35, 45, and 60 after healing. (D) Effect of KC cell sheet transplantation on scar area. (E) Immunohistochemical assay for α-SMA at the wound sites on day 28 after transplantation. Representative photomicrographs of α-SMA-positive cells were shown. (F) Immunohistochemical assay for α-SMA at the wound sites on day 28 after transplantation. Significantly fewer α-SMA-positive myofibroblasts were detected at the wound sites of the seq-915_x4024 group than those of the NC group. The number of cells was counted in visual fields under the microscope from five independent experiments (original magnification, ×200). (G) On day 28 after transplantation, the repaired tissues were stained with H&E. There were more-organized tissues, including skin appendages and new vessels, in the tissues of the seq-915_x4024 group compared with those in the tissues of the NC group. The matrix is uniformly arranged in the deep dermis of the seq-915_x4024 group. The results represent the means of the values. Bars indicate SD. *p < 0.05 and **p < 0.01, statistical significance between groups.
Figure 5.
KCs with Overexpressed Seq-915_x4024 Suppress Inflammatory Cell Infiltration and Scar Formation In Vivo
(A) On days 12 and 28 after transplantation, Masson’s Trichrome staining showing uniformly arranged collagen bundles in the deep dermis of the seq-915_x4024 group. (B) On days 1, 3, and 6 after transplantation, the tissues at the wound site were stained with H&E. The inflammatory cells were stained with blue nuclei, as indicated by black arrows. (C) Representative microscopic pictures of Ly6G immunofluorescence (green) from the tissues at the wound sites on days 1 and 3 after transplantation. There were significantly fewer Ly6G-positive neutrophils of the seq-915_x4024 group than in the NC group. (D) Representative microscopic images of F4/80-immunofluorescence (green) in the tissues at the wound sites on day 28 after transplantation. There were significantly fewer F4/80-positive macrophages in the seq-915_x4024 group than in the NC group.
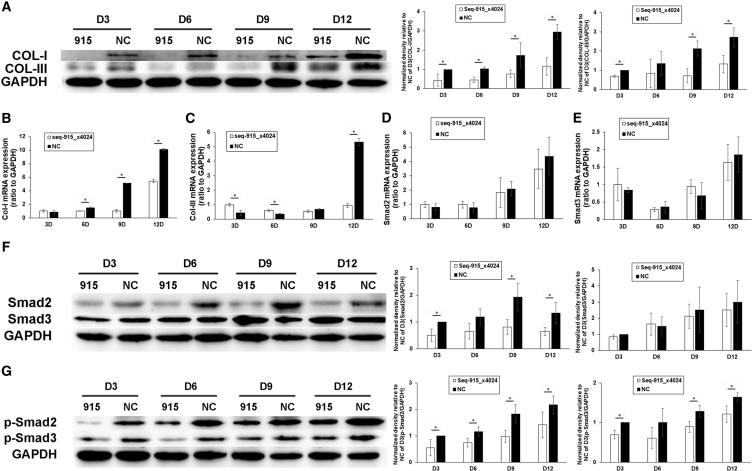
Seq-915_x4024 Suppresses the Expression of Col I, Col III, p-Smad2, and p-Smad3 In Vivo
On days 3, 6, 9, and 12 after transplantation, the wound tissues were harvested and analyzed using real-time qRT-PCR and western blot. Compared with the NC group, the protein expression of Col I and Col III and the mRNA expression of Col I were decreased significantly in the wounds of the seq-915_x4024 group (Figures 6A and 6B). On days 3 and 6 after transplantation, the mRNA expression of Col III in the wounds of the seq-915_x4024 group was higher than that in the NC group. However, on day 12, the mRNA expression of Col III in the NC group was significantly increased and higher than that in the seq-915_x4024 group (Figure 6C). Smad3 protein expression and mRNA expression of Smad2 and Smad3 were not significantly different between the seq-915_x4024 group and NC group (Figures 6D–6F). However, Smad2, p-Smad2, and p-Smad3 protein expression was significantly decreased in wounds of the seq-915_x4024 group compared with those of the NC group (Figures 6F and 6G).
Figure 6.
Seq-915_x4024 Suppresses the Expression of Col I, Col III, p-Smad2, and p-Smad3 In Vivo
On days 3, 6, 9, and 12 after transplantation, the wound tissues were harvested for further detection. (A) Western blotting was used to estimate the protein expression of Col I and Col III. (B) Real-time qRT-PCR was used to estimate the mRNA expression of Col I. (C) Real-time qRT-PCR was used to estimate the mRNA expression of Col III. (D) Real-time qRT-PCR was used to estimate the mRNA expression of Smad2. (E) Real-time qRT-PCR was used to estimate the mRNA expression of Smad3. (F) Western blotting was used to estimate the protein expression of Smad2 and Smad3. (G) Western blotting was used to estimate the protein expression of p-Smad2 and p-Smad3. The results represent the means of the values. Bars indicate SD. *p < 0.05, statistical significance between groups.
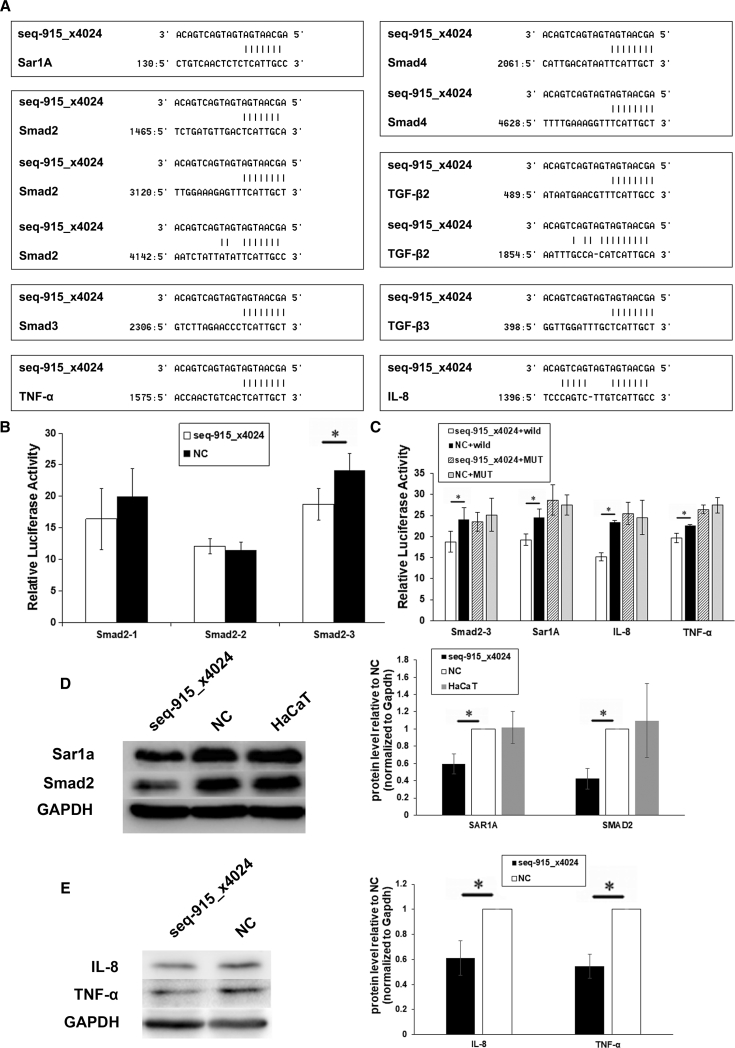
Seq-915_x4024 Targets Smad2, Sar1A, TNF-α, and IL-8
Using bioinformatics analyses, we found that TGF-β2, TGF-β3, Smad2, Smad3, Smad4, Sar1A, TNF-α, and IL-8 may be potential targets of seq-915_x4024 (Figure 7A). To confirm whether these genes are direct targets of seq-915_x4024, we performed a luciferase reporter assay. The results showed that the luciferase activity of pGL3-Sar1A-3′-UTR, pGL3-Smad2-3-3′-UTR, pGL3-TNF-α-3′-UTR, and pGL3-IL-8-3′-UTR reporters was significantly suppressed in seq-915_x4024-transfected HaCaT cells compared with the activity in NC-transfected HaCaT cells (Figures 7B and 7C). In contrast, there were no significant differences in the relative luciferase activity of pGL3-Smad2-3-MUT, pGL3-Sar1A-MUT, pGL3-TNF-α-MUT, or pGL3-IL-8-MUT reporters in seq-915_x4024-transfected HaCaT cells compared with activity in NC-transfected HaCaT cells (Figure 7C).
Figure 7.
Smad2, Sar1A, TNF-α, and IL-8 Are Directed Targets of Seq-915_x4024
(A) Potential target gene prediction of seq-915_x4024 using bioinformatics analyses. The 3′-UTRs of the genes were obtained from both the Ensembl and miRanda databases. (B) Analysis of luciferase activities of Smad2-1, -2, and -3 with seq-915_x4024 mimics or NCs in HaCaT cells. (C) Analysis of luciferase activities of Smad2-3, Smad2-3-MUT, Sar1A, Sar1A-MUT, TNF-α, TNF-α-MUT, IL-8, and IL-8-MUT with seq-915_x4024 mimics or NCs in HaCaT cells. (D) Protein expression analysis for Sar1A and Smad2 in transfected and parental HaCaT cells detected by western blotting. Normalized to an endogenous reference GAPDH, the level of the endogenous Sar1A and Smad2 showed a clear reduction in seq-915_x4024-transfected HaCaT cells. (E) Protein expression analysis of TNF-α and IL-8 in seq-915_x4024-transfected and NC-transfected HaCaT cells detected by western blotting. Normalized to GAPDH, the level of the endogenous TNF-α and IL-8 showed a clear reduction in seq-915_x4024-transfected HaCaT cells. The results represent the means of the values. Bars indicate SD. *p < 0.05, statistical significance between groups.
To further verify these results, we examined the Sar1A, Smad2, TNF-α, and IL-8 protein levels in seq-915_x4024-transfected, NC-transfected, and parental HaCaT cells by western blot. Normalized to the endogenous reference GAPDH, the levels of the endogenous Sar1A, Smad2, TNF-α, and IL-8 showed a clear reduction in seq-915_x4024-transfected HaCaT cells (Figures 7D and 7E). These results demonstrated that seq-915_x4024 may target Sar1A, Smad2, TNF-α, and IL-8 in KCs.
Discussion
It is beginning to be revealed that miRNAs play important roles in skin regeneration.15 In early to mid-gestation, several miRNAs, especially skin-specific expressed miRNAs, contribute to skin regeneration without scar formation by targeting key genes and pathways.16, 17 Using next-generation sequencing and mirTools 2.0,18, 19 we identified a novel miRNA candidate, seq-915_x4024, which was highly expressed in mid-gestational fetal KCs. In this study, we further investigated the role of seq-915_x4024 in the process of wound healing.
Early in gestation, wounds heal rapidly. In addition, fetal KCs exhibit a significantly increased proliferation rate and promote the migration and proliferation ability of FBs compared with late-gestational fetal KCs.11, 12 Are these fetal KC cell activities associated with overexpression of seq-915_x4024? We demonstrated that KCs with overexpression of seq-915_x4024 exhibit the same biological behaviors as fetal KCs at mid-gestation. After being transfected with seq-915_x4024 mimics, HaCaT cells exhibited higher proliferative activity and the ability to promote FB migration and FB proliferation.
To investigate whether overexpression of seq-915_x4024 in KCs plays a role in the process of wound repair, an in vivo wound-healing model was used. We demonstrated that wounds in the seq-915_x4024 group heal with fewer myofibroblasts, less scar tissue, uniformly arranged collagen bundles in the deep dermis, more-organized tissue, and less expression of Col I and Col III. Collagens are the most important components of dermal ECM. The key features of fibrosis are increased collagen content, and the arrangement of the collagen fibers becomes disordered. The accumulation of myofibroblasts is also involved in the process of fibrosis because myofibroblasts produce a collagen-rich matrix that forms scar tissues.20 This evidence demonstrates that seq-915_x4024 significantly inhibits FB conversion into myofibroblasts and scar formation.
Fetal skin synthesizes less Col I and more Col III than adult skin.21 Compared with type I collagen fibers, type III collagen fibers are smaller and may be deposited in a well-structured reticular manner that is similar to the structure of the fibers present in normal skin.22, 23 In this study, an increased ratio of type III versus type I collagen was found in seq-915_x4024-transfected HaCaT cells, indicating that seq-915_x4024 contributes to the collagen composition, which leads to skin regeneration. In addition, we found that Has3 was also upregulated by seq-915_x4024. Has3, one of the three types of Has, synthesizes hyaluronic acid, which plays major roles in scarless wound healing.24 In dermal wounds receiving hyaluronic acid treatment, scar formation is significantly reduced.25, 26, 27, 28 These results suggest that seq-915_x4024 regulates ECM components, including Col I, Col III, and Has3, which may contribute to skin regeneration.
Inflammation is a key determinant of fibrosis. The inflammatory phase is the beginning of wound healing. During this phase, chemokines and cytokines, such as platelet-derived growth factor (PDGF), interleukins, TGF-β, and TNF-α, are released into the bloodstream and activate inflammatory cells.29, 30, 31 Early in gestation, wounds in fetal mammalian skin do not trigger inflammation in response to damage and heal without scarring. In contrast, fetal wounds generated during late gestation heal with a strong inflammatory response and scar formation.32
Chemokines and cytokines play important roles in scarless wound healing in fetal skin. Inflammatory cells exist and respond to inflammatory factors, even at the stage when fetal cutaneous wounds heal without an inflammatory response.33, 34 TNF-α is an important pro-inflammatory cytokine. After wounding, in response to TNF-α, CXC family members, particularly CXCLs 1, 5, and 8, are released to recruit neutrophils into inflamed tissues.35, 36, 37 To investigate whether overexpression of seq-915_x4024 in KCs plays an anti-inflammatory role in wound healing, we used TNF-α as an inflammatory stimulus38 to demonstrate that the expression levels of IL-6, IL-8, CXCL1, CXCL5, and TNF-α are significantly suppressed in KCs transfected with seq-915_x4024. TNF-α and IL-8 are direct targets of seq-915_x4024 in KCs. IL-6 and IL-8 (pro-inflammatory cytokines) are expressed at low levels in fetal wounds.39, 40 Attenuating IL-6 and IL-8 expression leads to reduced recruitment of inflammatory cells and decreased scar formation.41 Using in vivo experiments, we demonstrated that KC cell sheets with overexpressed seq-915_x4024 suppress inflammatory cell infiltration. We believe that seq-915_x4024 also functions as an anti-inflammatory factor and contributes to skin regeneration by suppressing the pro-inflammatory cytokines TNF-α, IL-6, and IL-8 and the pro-inflammatory chemokines CXCL1 and CXCL5.
Among all the growth factors involved in wound healing, TGF-β family members are the most important factors that play roles in all three phases, especially in the remodeling phase. The mammalian genome contains three TGF-β isoforms known as TGF-β1, TGF-β2, and TGF-β3. TGF-β1 is well known as a key mediator in tissue fibrosis via the Smad signaling pathway. The activated TGF-β1-Smad signaling pathway induces myofibroblastic differentiation, induces FB production of type I collagen, and promotes ECM deposition.42, 43 In contrast, TGF-β3 appears to play different roles in wound healing, as it inhibits myofibroblast proliferation and type I collagen deposition, which leads to scar tissue formation.44 Overexpression of seq-915_x4024 leading to higher TGF-β3 and lower TGF-β1 may contribute to suppressing fibrosis.
What is the molecular mechanism by which seq-915_x4024 acts as an anti-fibrotic factor? TGF-β1 plays a central role in tissue fibrosis via the Smad signaling pathway.45 TGF-β1 interacting with type I and type II transmembrane receptors (TGF-βRI and TGF-RII, respectively) activates canonical Smad pathway members, including Sar1A (Smad anchor for receptor activation, SARA) and the Smad family.46 Upregulating the expression of TGF-β1 stimulates overabundant collagen synthesis by activating the phosphorylation of Smad2 and Smad3, forming hetero-oligomeric complexes with Smad4 (common Smads, co-Smads), and finally translocating into the nucleus and activating transcription.47 We investigated whether TGF-β1-Smad signaling pathway members are targets of seq-915_x4024 and demonstrated that Sar1A and Smad2 are direct targets. In addition, the expression of p-Smad2 and p-Smad3 was significantly decreased by seq-915_x4024 in vivo. These results demonstrated that seq-915_x4024 suppresses the TGF-β1-Smad signaling pathway by targeting Sar1A and Smad2. Seq-915_x4024 acts as an anti-fibrotic factor by suppressing the TGF-β1-Smad signaling pathway members Sar1A and Smad2.
In conclusion, our study shows that the overexpression of seq-915_x4024 in KCs suppresses inflammatory cell infiltration and scar formation by targeting the TGF-β1-Smad signaling pathway and inflammatory factors TNF-α and IL-8. Seq-915_x4024 could be a new anti-fibrotic factor for the treatment of wound healing.
Materials and Methods
Skin Samples and Cell Culture
The immortalized human KC cell line HaCaT was kindly provided by Chundi He and propagated in high-glucose Dulbecco’s modified Eagle’s medium (DMEM [HG]; Invitrogen Life Technologies, Carlsbad, CA, USA). Skin samples were obtained from First Hospital of China Medical University. This study was performed in accordance with the ARRIVE guidelines and under a research protocol approved by the Ethics Committee of China Medical University. Written informed consent was obtained.
Primary culture of FBs was obtained from skin samples. Cells were prepared as previously described.48 Briefly, full-thickness skin samples were de-epithelialized after incubating at 4°C overnight in Dispase II (Roche Applied Science, Indianapolis, IN, USA). After mincing the dermal components, the isolated FBs were cultured in DMEM (HG) supplemented with 10% FBS (Invitrogen Life Technologies), 100 U/mL penicillin and 100 μg/mL streptomycin. The cultures were incubated at 37°C in a humidified incubator with 5% CO2. When the FBs reached 80%–90% confluence, the cells were passaged for expansion. For the experiments, human dermal FBs were used at passages 3–6.
RNA Isolation and Real-Time qRT-PCR
Total RNA was extracted using the mirVana miRNA Isolation Kit (Thermo Fisher, Uppsala, Sweden) according to the manufacturer’s instructions. The concentration and purity of RNA were determined by ultraviolet spectrophotometry (A260/A280 > 1.9), using a nano-photometer UV-visible spectrophotometer (Implen, Schatzbogen, Germany). The 3′-termini of the RNA segments were polyadenylated using the Poly(A) Tailing Kit (Thermo Fisher). Then, RNA was extracted with phenol-chloroform and precipitated with ethanol.
Real-time qRT-PCR was used to confirm the expression levels of seq-915_x4024, IL-1α, IL-1β, IL-6, IL-8, TNF-α, CXCL1, CXCL5, TGF-β1, TGF-β3, Has3, Col I, and Col III. Reverse transcription was performed using a PrimeScript RT reagent Kit with gDNA Eraser (Takara Bio, Shiga, Japan). Real-time qRT-PCR was performed on a 7500 Real-Time PCR system (Applied Biosystems, Foster City, CA, USA) supplied with analytical software, using an SYBR Premix Ex Taq II Kit (Takara Bio) according to the manufacturer’s instructions. The U6 or GAPDH mRNA level, as an endogenous reference, was used for normalization. The relative expression levels of the genes were calculated using the 2–ΔΔCT equation in which ΔCT = CT miRNA − CT U6 or ΔCT = CT mRNA − CT GAPDH.49 The primers used for real-time qRT-PCR are given in Table S1.
Transfection
All RNA oligoribonucleotides and their respective NCs were purchased from GenePharma (Shanghai, China). The pyrimidine nucleotides in the seq-915_x4024 mimics, anti-seq-915_x4024, and their respective NCs were substituted with their 2-O-methyl analogs to improve RNA stability. HaCaT cells were plated 1 day before transfection. RNA oligoribonucleotides were transfected using Lipofectamine 2000 (Thermo Fisher) according to the manufacturer’s instructions. After 48 h, the cells were harvested for further experiments.
Coculture of Dermal FBs with HaCaT
Dermal FBs were cocultured with parental and transfected HaCaT cells in Transwell chambers (3-μm pore size; Corning, Corning, NY, USA). After 96 h, FBs were harvested for further experiments.
Transwell Cell Migration Assay
We used a Transwell cell migration assay to estimate the migration ability of the cells in vitro. Before use, the bottom of the culture inserts (8-μm pores) in 24-well tissue culture plates (Transwell; Corning) was coated with serum-free medium at 37°C for 1 h. A total of 5 × 104 cells were harvested by trypsinization, washed with serum-free medium, resuspended at a concentration of 5 × 105/mL, and placed in the upper chamber. The lower chamber contained 10% FBS for use as a chemoattractant. After 24 h of incubation at 37°C with 5% CO2, the number of cells that had migrated to the basal side of the membrane was quantified by counting 16 independent symmetrical visual fields under the microscope, and cell morphology was observed by staining with H&E (hematoxylin-eosin).
Quantification of Cell Proliferation
Cell proliferation was determined using a colorimetric microculture assay with MTS (3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium) dye. A total of 5 × 103 HaCaT cells or 2 × 103 FBs were seeded separately into 96-well culture plates for 24, 48, 72, and 96 h; after incubation with 20 μL MTS for 2 h at 37°C, the absorbance was measured using an iMARK microplate reader (Bio-Rad, Hercules, CA, USA) at a wavelength of 495 nm.
In Vitro Wound-Healing Assay
For the migration assay, culture inserts (Ibidi, Munich, Germany) were placed into the wells of a six-well plate. After being cocultured, 5 × 103 FBs with 100 μL medium were seeded on each side of a culture insert and incubated under normal culture conditions to allow the cells to grow to confluence. Afterward, inserts were removed, leaving a cell-free area. The medium was switched to 2 mL DMEM (HG) supplemented with 1% FBS. The status of the scratch wounds was monitored using an inverted microscope at 0, 12, 24, and 48 h, and representative images were collected. The results are presented as the percentage of wound healing, calculated as follows: [wound area (initial) − wound area (final)]/wound area (initial) × 100.
KC-Sheet Preparation and Wound-Healing Model In Vivo
HaCaT cells (5 × 106) were seeded into Nunc UpCell (Thermo Fisher) dishes and incubated under normal culture conditions to allow cells to grow to confluence. After 24 h, the dishes were transferred to another incubator, set at 20°C with 5% CO2, for 4 h to release the cultured cells as a KC cell sheet.
In vivo assessment of the effect of seq-915_x4024 on cutaneous regeneration was performed using a wound-healing model. Female Wistar rats (10–12 weeks old, approximately 250 g) were obtained from Vital River Laboratory Animal Technology (Beijing, China) and housed in the animal care facilities of China Medical University under specific-pathogen-free conditions. After hair removal from the dorsal surface, the rats were anesthetized with pentobarbital sodium (40 mg/kg; Solarbio, China). Full-thickness skin, 10 mm in diameter, was removed from each side of the dorsal skin of each rat. The seq-915_x4024-transfected KC cell sheet was transplanted onto the surface of the left-hand wound bed, and the NC-transfected KC cell sheet was transplanted on the other side as an NC, followed by coating with Hydrosorb (Hartmann, Germany), Tegaderm film dressing (3M Health Care, Neuss, Germany), and medical gauze. After rats were sacrificed, the wound tissues, including a border of normal tissue, were harvested and immediately fixed in 4% paraformaldehyde in phosphate-buffered saline and were dehydrated in a 30% sucrose solution. Frozen sections were prepared in a freezing microtome (Leica, Germany) and subjected to the following procedures: (1) H&E staining for histological evaluation; (2) Masson’s trichrome staining (Solarbio, Beijing, China) for collagen fibers; (3) immunohistochemical assay for the expression of α-SMA (α-smooth muscle actin); and (4) immunofluorescence assay for expression of Ly6G and F4/80.
Bioinformatics Analyses and Luciferase Reporter Assays
For potential target gene prediction of seq-915_x4024, the 3′-UTRs of the genes were obtained from both the Ensembl and miRanda databases. We considered the seed sequence (nucleotides 2–8) of seq-915_x4024 and at least one part of the 3′-UTR of target mRNAs was reverse complemented perfectly.10
For luciferase reporter experiments, the wild-type 3′-UTR segments of Sar1A, Smad2, Smad3, Smad4, TGF-β2, TGF-β3, TNF-α, and IL-8, containing the seq-915_x4024 binding sites, were amplified by PCR and inserted into the pGL3-control vector (Promega, Madison, WI, USA) using the XbaI site, which is immediately downstream of the luciferase stop codon. DNA segments with scrambled target sites (Smad2-3-MUT, Sar1A-MUT, TNF-α-MUT, and IL-8-MUT) designed to interfere with seed sequence recognition were also cloned to serve as controls. HaCaT cells were plated in 24-well plates. For each well, 20 pM seq-915_x4024 mimics or NC, 0.8 μg of the firefly luciferase report vector and 0.08 μg of the control vector containing Renilla luciferase, pRL-TK (Promega), were transfected using Lipofectamine 2000. Twenty-four hours after transfection, Firefly and Renilla luciferase activities were measured consecutively using a dual-luciferase reporter assay (Promega) on a Centro LB 960 (Berthold, Bad Wildbad, Germany).
Western Blot Analysis
Western blotting was performed as previously described.13 Briefly, total protein was extracted using a Total Protein Extraction Kit (KeyGen, Nanjing, China), followed by measurement using a BCA Protein Assay Kit (KeyGen). A total of 30 μg of each sample was separated by 12% SDS polyacrylamide gels and electrophoretically transferred to polyvinylidene difluoride membranes (Millipore, Billerica, MA, USA). After incubation with 5% bovine serum albumin in Tris-buffered saline-0.5% Tween-20 at room temperature for 1 h, the membranes were incubated at 4°C overnight with primary antibodies against IL-6 (ImmunoWay, Plano, TX, USA), IL-8 (ImmunoWay), TNF-α (ImmunoWay), Sar1A (ImmunoWay), Smad2 (ImmunoWay), TGF-β1 (Abcam, Cambridge, MA, USA), TGF-β3 (Santa Cruz Biotechnology, Santa Cruz, CA, USA), Has3 (Abcam), Col III (ImmunoWay), and GAPDH (Santa Cruz) and then incubated with horseradish peroxidase-conjugated secondary antibody at room temperature for 2 h. The antigen-antibody complexes were visualized, using an ECL Kit (Pierce, Rockford, IL, USA). Quantification of protein was carried out using FluorChem 2.01 (Alpha Innotech, San Leandro, CA, USA). Protein levels in seq-915_x4024-transfected HaCaT cells were presented as fold change normalized to an endogenous reference (GAPDH protein) and relative to NC-transfected HaCaT cells.
Statistical Analysis
The results are presented as the mean ± SD from at least three separate experiments. Statistical differences between groups were analyzed using a one-way ANOVA test or Student’s t test. The statistical analyses were performed using SPSS 16.0 software (SPSS, Chicago, IL, USA). Values of p less than 0.05 were considered statistically significant.
Author Contributions
F.Z. and H.L. performed the experiments; F.Z. wrote the paper; Z.W., X.L., and D.Z. provided study materials; T.Z., R.W., and X.L. contributed to data analysis and interpretation; P.S. contributed to collection and assembly of data; and X.P. designed the study and provided final approval of the manuscript.
Conflicts of Interest
The authors declare no competing interests.
Acknowledgments
This work was supported by the Program for National Basic Research Program of China (2012CB518103), the National Science Foundation of China (81370883), the National Key Research and Development Project (2017TFC1103300), the Shenyang Key Laboratory Project (F15-157-1-00), the Key R & D and Technology Transfer Program (Z17-5-039), and the New Teacher Foundation of China Medical University (XZR 20160023).
Footnotes
Supplemental Information includes one figure and one table and can be found with this article online at https://doi.org/10.1016/j.omtn.2018.12.016.
Supplemental Information
References
- 1.Bayer A., Tohidnezhad M., Lammel J., Lippross S., Behrendt P., Klüter T., Pufe T., Jahr H., Cremer J., Rademacher F. Platelet-released growth factors induce differentiation of primary keratinocytes. Mediators Inflamm. 2017;2017:5671615. doi: 10.1155/2017/5671615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yang X., Wang J., Guo S.L., Fan K.J., Li J., Wang Y.L., Teng Y., Yang X. miR-21 promotes keratinocyte migration and re-epithelialization during wound healing. Int. J. Biol. Sci. 2011;7:685–690. doi: 10.7150/ijbs.7.685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jin Y., Tymen S.D., Chen D., Fang Z.J., Zhao Y., Dragas D., Dai Y., Marucha P.T., Zhou X. MicroRNA-99 family targets AKT/mTOR signaling pathway in dermal wound healing. PLoS ONE. 2013;8:e64434. doi: 10.1371/journal.pone.0064434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Seo G.Y., Lim Y., Koh D., Huh J.S., Hyun C., Kim Y.M., Cho M. TMF and glycitin act synergistically on keratinocytes and fibroblasts to promote wound healing and anti-scarring activity. Exp. Mol. Med. 2017;49:e302. doi: 10.1038/emm.2016.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ho J., Walsh C., Yue D., Dardik A., Cheema U. Current advancements and strategies in tissue engineering for wound healing: a comprehensive review. Adv. Wound Care (New Rochelle) 2017;6:191–209. doi: 10.1089/wound.2016.0723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huen A.C., Marathi A., Nam P.K., Wells A. CXCL11 expression by keratinocytes occurs transiently between reaching confluence and cellular compaction. Adv. Wound Care (New Rochelle) 2016;5:517–526. doi: 10.1089/wound.2015.0680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen W., Fu X., Ge S., Sun T., Zhou G., Jiang D., Sheng Z. Ontogeny of expression of transforming growth factor-beta and its receptors and their possible relationship with scarless healing in human fetal skin. Wound Repair Regen. 2005;13:68–75. doi: 10.1111/j.1067-1927.2005.130109.x. [DOI] [PubMed] [Google Scholar]
- 8.Samuels P., Tan A.K. Fetal scarless wound healing. J. Otolaryngol. 1999;28:296–302. [PubMed] [Google Scholar]
- 9.Dang C., Ting K., Soo C., Longaker M.T., Lorenz H.P. Fetal wound healing current perspectives. Clin. Plast. Surg. 2003;30:13–23. doi: 10.1016/s0094-1298(02)00067-6. [DOI] [PubMed] [Google Scholar]
- 10.Zhao F., Wang Z., Lang H., Liu X., Zhang D., Wang X., Zhang T., Wang R., Shi P., Pang X. Dynamic expression of novel MiRNA candidates and MiRNA-34 family members in early- to mid-gestational fetal keratinocytes contributes to scarless wound healing by targeting the TGF-β pathway. PLoS ONE. 2015;10:e0126087. doi: 10.1371/journal.pone.0126087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang Z., Liu X., Zhang D., Wang X., Zhao F., Zhang T., Wang R., Lin X., Shi P., Pang X. Phenotypic and functional modulation of 20-30 year old dermal fibroblasts by mid- and late-gestational keratinocytes in vitro. Burns. 2015;41:1064–1075. doi: 10.1016/j.burns.2014.12.013. [DOI] [PubMed] [Google Scholar]
- 12.Wang Z., Liu X., Zhang D., Wang X., Zhao F., Shi P., Pang X. Co-culture with human fetal epidermal keratinocytes promotes proliferation and migration of human fetal and adult dermal fibroblasts. Mol. Med. Rep. 2015;11:1105–1110. doi: 10.3892/mmr.2014.2798. [DOI] [PubMed] [Google Scholar]
- 13.Xu Y., Zhao F., Wang Z., Song Y., Luo Y., Zhang X., Jiang L., Sun Z., Miao Z., Xu H. MicroRNA-335 acts as a metastasis suppressor in gastric cancer by targeting Bcl-w and specificity protein 1. Oncogene. 2012;31:1398–1407. doi: 10.1038/onc.2011.340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dong X., Zhang C., Ma S., Wen H. Mast cell chymase in keloid induces profibrotic response via transforming growth factor-β1/Smad activation in keloid fibroblasts. Int. J. Clin. Exp. Pathol. 2014;7:3596–3607. [PMC free article] [PubMed] [Google Scholar]
- 15.Horsburgh S., Fullard N., Roger M., Degnan A., Todryk S., Przyborski S., O’Reilly S. MicroRNAs in the skin: role in development, homoeostasis and regeneration. Clin. Sci. (Lond.) 2017;131:1923–1940. doi: 10.1042/CS20170039. [DOI] [PubMed] [Google Scholar]
- 16.Cheng J., Yu H., Deng S., Shen G. MicroRNA profiling in mid- and late-gestational fetal skin: implication for scarless wound healing. Tohoku J. Exp. Med. 2010;221:203–209. doi: 10.1620/tjem.221.203. [DOI] [PubMed] [Google Scholar]
- 17.Kathju S., Gallo P.H., Satish L. Scarless integumentary wound healing in the mammalian fetus: molecular basis and therapeutic implications. Birth Defects Res. C Embryo Today. 2012;96:223–236. doi: 10.1002/bdrc.21015. [DOI] [PubMed] [Google Scholar]
- 18.Jung C.H., Hansen M.A., Makunin I.V., Korbie D.J., Mattick J.S. Identification of novel non-coding RNAs using profiles of short sequence reads from next generation sequencing data. BMC Genomics. 2010;11:77. doi: 10.1186/1471-2164-11-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wu J., Liu Q., Wang X., Zheng J., Wang T., You M., Sheng Sun Z., Shi Q. mirTools 2.0 for non-coding RNA discovery, profiling, and functional annotation based on high-throughput sequencing. RNA Biol. 2013;10:1087–1092. doi: 10.4161/rna.25193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wise L.M., Stuart G.S., Real N.C., Fleming S.B., Mercer A.A. VEGF receptor-2 activation mediated by VEGF-E limits scar tissue formation following cutaneous injury. Adv. Wound Care (New Rochelle) 2018;7:283–297. doi: 10.1089/wound.2016.0721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Leung A., Crombleholme T.M., Keswani S.G. Fetal wound healing: implications for minimal scar formation. Curr. Opin. Pediatr. 2012;24:371–378. doi: 10.1097/MOP.0b013e3283535790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zgheib C., Xu J., Liechty K.W. Targeting inflammatory cytokines and extracellular matrix composition to promote wound regeneration. Adv. Wound Care (New Rochelle) 2014;3:344–355. doi: 10.1089/wound.2013.0456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Merkel J.R., DiPaolo B.R., Hallock G.G., Rice D.C. Type I and type III collagen content of healing wounds in fetal and adult rats. Proc. Soc. Exp. Biol. Med. 1988;187:493–497. doi: 10.3181/00379727-187-42694. [DOI] [PubMed] [Google Scholar]
- 24.Carre A.L., James A.W., MacLeod L., Kong W., Kawai K., Longaker M.T., Lorenz H.P. Interaction of wingless protein (Wnt), transforming growth factor-beta1, and hyaluronan production in fetal and postnatal fibroblasts. Plast. Reconstr. Surg. 2010;125:74–88. doi: 10.1097/PRS.0b013e3181c495d1. [DOI] [PubMed] [Google Scholar]
- 25.Akershoek J.J., Vlig M., Talhout W., Boekema B.K., Richters C.D., Beelen R.H., Brouwer K.M., Middelkoop E., Ulrich M.M. Cell therapy for full-thickness wounds: are fetal dermal cells a potential source? Cell Tissue Res. 2016;364:83–94. doi: 10.1007/s00441-015-2293-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Faga A., Nicoletti G., Brenta F., Scevola S., Abatangelo G., Brun P. Hyaluronic acid three-dimensional scaffold for surgical revision of retracting scars: a human experimental study. Int. Wound J. 2013;10:329–335. doi: 10.1111/j.1742-481X.2012.00981.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mahedia M., Shah N., Amirlak B. Clinical evaluation of hyaluronic acid sponge with zinc versus placebo for scar reduction after breast surgery. Plast. Reconstr. Surg. Glob. Open. 2016;4:e791. doi: 10.1097/GOX.0000000000000747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hussain S.N., Goodman G.J., Rahman E. Treatment of a traumatic atrophic depressed scar with hyaluronic acid fillers: a case report. Clin. Cosmet. Investig. Dermatol. 2017;10:285–287. doi: 10.2147/CCID.S132626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fahs F., Bi X., Yu F.S., Zhou L., Mi Q.S. New insights into microRNAs in skin wound healing. IUBMB Life. 2015;67:889–896. doi: 10.1002/iub.1449. [DOI] [PubMed] [Google Scholar]
- 30.O’Reilly S. MicroRNAs in fibrosis: opportunities and challenges. Arthritis Res. Ther. 2016;18:11. doi: 10.1186/s13075-016-0929-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.O’Reilly S. Epigenetics in fibrosis. Mol. Aspects Med. 2017;54:89–102. doi: 10.1016/j.mam.2016.10.001. [DOI] [PubMed] [Google Scholar]
- 32.Dardenne A.D., Wulff B.C., Wilgus T.A. The alarmin HMGB-1 influences healing outcomes in fetal skin wounds. Wound Repair Regen. 2013;21:282–291. doi: 10.1111/wrr.12028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cowin A.J., Brosnan M.P., Holmes T.M., Ferguson M.W. Endogenous inflammatory response to dermal wound healing in the fetal and adult mouse. Dev. Dyn. 1998;212:385–393. doi: 10.1002/(SICI)1097-0177(199807)212:3<385::AID-AJA6>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- 34.Kishi K., Okabe K., Shimizu R., Kubota Y. Fetal skin possesses the ability to regenerate completely: complete regeneration of skin. Keio J. Med. 2012;61:101–108. doi: 10.2302/kjm.2011-0002-ir. [DOI] [PubMed] [Google Scholar]
- 35.Charo I.F., Ransohoff R.M. The many roles of chemokines and chemokine receptors in inflammation. N. Engl. J. Med. 2006;354:610–621. doi: 10.1056/NEJMra052723. [DOI] [PubMed] [Google Scholar]
- 36.Zaja-Milatovic S., Richmond A. CXC chemokines and their receptors: a case for a significant biological role in cutaneous wound healing. Histol. Histopathol. 2008;23:1399–1407. doi: 10.14670/hh-23.1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rees P.A., Greaves N.S., Baguneid M., Bayat A. Chemokines in wound healing and as potential therapeutic targets for reducing cutaneous scarring. Adv. Wound Care (New Rochelle) 2015;4:687–703. doi: 10.1089/wound.2014.0568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schneider M., Efferth T., Abdel-Aziz H. Anti-inflammatory effects of herbal preparations STW5 and STW5-II in cytokine-challenged normal human colon cells. Front. Pharmacol. 2016;7:393. doi: 10.3389/fphar.2016.00393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liechty K.W., Adzick N.S., Crombleholme T.M. Diminished interleukin 6 (IL-6) production during scarless human fetal wound repair. Cytokine. 2000;12:671–676. doi: 10.1006/cyto.1999.0598. [DOI] [PubMed] [Google Scholar]
- 40.Liechty K.W., Crombleholme T.M., Cass D.L., Martin B., Adzick N.S. Diminished interleukin-8 (IL-8) production in the fetal wound healing response. J. Surg. Res. 1998;77:80–84. doi: 10.1006/jsre.1998.5345. [DOI] [PubMed] [Google Scholar]
- 41.Morris M.W., Jr., Allukian M., 3rd, Herdrich B.J., Caskey R.C., Zgheib C., Xu J., Dorsett-Martin W., Mitchell M.E., Liechty K.W. Modulation of the inflammatory response by increasing fetal wound size or interleukin-10 overexpression determines wound phenotype and scar formation. Wound Repair Regen. 2014;22:406–414. doi: 10.1111/wrr.12180. [DOI] [PubMed] [Google Scholar]
- 42.Penn J.W., Grobbelaar A.O., Rolfe K.J. The role of the TGF-β family in wound healing, burns and scarring: a review. Int. J. Burns Trauma. 2012;2:18–28. [PMC free article] [PubMed] [Google Scholar]
- 43.Vallée A., Lecarpentier Y., Guillevin R., Vallée J.N. Interactions between TGF-β1, canonical WNT/β-catenin pathway and PPAR γ in radiation-induced fibrosis. Oncotarget. 2017;8:90579–90604. doi: 10.18632/oncotarget.21234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li M., Qiu L., Hu W., Deng X., Xu H., Cao Y., Xiao Z., Peng L., Johnson S., Alexey L. Genetically-modified bone mesenchymal stem cells with TGF-β3 improve wound healing and reduce scar tissue formation in a rabbit model. Exp. Cell Res. 2018;367:24–29. doi: 10.1016/j.yexcr.2018.02.006. [DOI] [PubMed] [Google Scholar]
- 45.Verrecchia F., Mauviel A. Transforming growth factor-beta and fibrosis. World J. Gastroenterol. 2007;13:3056–3062. doi: 10.3748/wjg.v13.i22.3056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Guo X., Hutcheon A.E.K., Tran J.A., Zieske J.D. TGF-β-target genes are differentially regulated in corneal epithelial cells and fibroblasts. New Front. Ophthalmol. 2017;3 doi: 10.15761/NFO.1000151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lee W.J., Choi I.K., Lee J.H., Lee J.S., Kim Y.O., Rah D.K., Yun C.O. Relaxin-expressing adenovirus decreases collagen synthesis and up-regulates matrix metalloproteinase expression in keloid fibroblasts: in vitro experiments. Plast. Reconstr. Surg. 2012;130:407e–417e. doi: 10.1097/PRS.0b013e31825dbf56. [DOI] [PubMed] [Google Scholar]
- 48.Liu X., Wang Z., Wang R., Zhao F., Shi P., Jiang Y., Pang X. Direct comparison of the potency of human mesenchymal stem cells derived from amnion tissue, bone marrow and adipose tissue at inducing dermal fibroblast responses to cutaneous wounds. Int. J. Mol. Med. 2013;31:407–415. doi: 10.3892/ijmm.2012.1199. [DOI] [PubMed] [Google Scholar]
- 49.Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−delta delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.