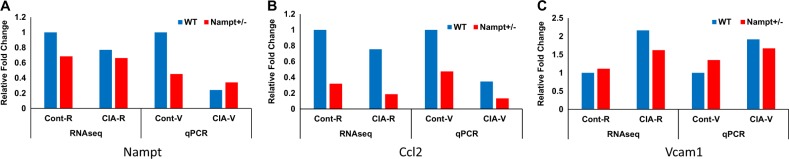
Fig. 6. Validation of differential gene expression profiles determined by RNA seq via TaqMan™ qPCR analyses of selected targets.
RNA isolation, RNA-seq, and RT-PCR were carried out as described in Materials and methods. All results of gene expression levels are expressed as relative fold changes. The relative fold changes represent mean values of technical replicates of N = 6 and biological replicates of N = 2. a Comparison of relative changes in Nampt expression levels for RNA-seq and qPCR. b Relative fold changes for Ccl2. c Relative fold changes Vcam1. All fold changes are relative to the wild-type (WT) control. The ∆∆CT was calculated by subtracting the mean ∆CT for the WT control group from the mean ∆CT for each experimental sample. Gapdh served as the endogenous control. Cont-R control RNA seq, CIA-R CIA RNA seq, Cont-V control TaqMan™ validation, CIA-V CIA TaqMan™ validation