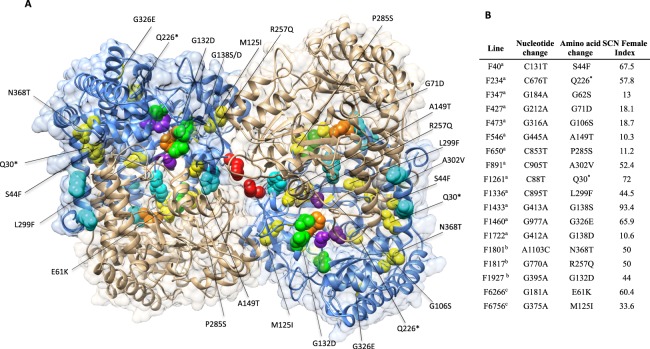
Figure 7.
The identified eighteen EMS Gmshmt08c mutants. Homology modeling of a GmSHMT08c asymmetric homotetramer predicted protein structure, showing all the eighteen identified Gmshmt08c mutants, in addition to important GmSHMT08c residues. Four subunits are shown, representing a GmSHMT08c homomer each; (A) GmSHMT08 homomer upper right, (B) GmSHMT08 homomer down right, (C) GmSHMT08 homomer upper left, (D) GmSHMT08 homomer down left. Only GmSHMT08c subunits (B,C) were highlighted with PLP/catalytic sites (orange), PLP cofactor binding (purple), THF cofactor binding (green), oligomeric structure maintenance (gray), dimerization (cyan), and tetramerization (red) residues. All eighteen Gmshmt08c mutants were mapped, represented in yellow sphere, and labeled in the subunits (B,C). All the susceptible EMS mutagenized Forrest mutants that were identified by forward genetics presented mutations at the GmSHMT08c gene only. aMutants identified by forward genetics (Kandoth et al.40). bMutants identified by forward genetics in the current study, cMutants identified by TILLING (Liu et al.3).