Sediments sequester organic matter over geologic time scales and impact global climate regulation. Microbial communities in marine sediments drive organic matter degradation, but the factors controlling their assemblages and activities, which in turn impact their role in organic matter degradation, are not well understood. Hence, determining the role of microbial communities in carbon cycling in various sediment types is necessary for predicting future sediment carbon cycling. We examined microbial communities in Baltic Sea sediments, which were deposited across various climatic and geographical regimes to determine the relationship between microbial potential for breakdown of organic matter and abiotic factors, including geochemistry and sediment lithology. The findings from this study will contribute to our understanding of carbon cycling in the deep biosphere and how microbial communities live in deeply buried environments.
KEYWORDS: Baltic Sea, heterotrophy, microbial ecology, sediment
ABSTRACT
Globally, marine sediments are a vast repository of organic matter, which is degraded through various microbial pathways, including polymer hydrolysis and monomer fermentation. The sources, abundances, and quality (i.e., labile or recalcitrant) of the organic matter and the composition of the microbial assemblages vary between sediments. Here, we examine new and previously published sediment metagenomes from the Baltic Sea and the nearby Kattegat region to determine connections between geochemistry and the community potential to degrade organic carbon. Diverse organic matter hydrolysis encoding genes were present in sediments between 0.25 and 67 meters below seafloor and were in higher relative abundances in those sediments that contained more organic matter. New analysis of previously published metatranscriptomes demonstrated that many of these genes were transcribed in two organic-rich Holocene sediments. Some of the variation in deduced pathways in the metagenomes correlated with carbon content and depositional conditions. Fermentation-related genes were found in all samples and encoded multiple fermentation pathways. Notably, genes involved in alcohol metabolism were amongst the most abundant of these genes, indicating that this is an important but underappreciated aspect of sediment carbon cycling. This study is a step towards a more complete understanding of microbial food webs and the impacts of depositional facies on present sedimentary microbial communities.
IMPORTANCE Sediments sequester organic matter over geologic time scales and impact global climate regulation. Microbial communities in marine sediments drive organic matter degradation, but the factors controlling their assemblages and activities, which in turn impact their role in organic matter degradation, are not well understood. Hence, determining the role of microbial communities in carbon cycling in various sediment types is necessary for predicting future sediment carbon cycling. We examined microbial communities in Baltic Sea sediments, which were deposited across various climatic and geographical regimes to determine the relationship between microbial potential for breakdown of organic matter and abiotic factors, including geochemistry and sediment lithology. The findings from this study will contribute to our understanding of carbon cycling in the deep biosphere and how microbial communities live in deeply buried environments.
INTRODUCTION
Organic matter (OM) burial in marine sediments sequesters carbon over geologic time and thereby plays a role in climate regulation. Globally, marine sediments store 7.8 × 1022 g of carbon, including organic matter from both terrestrial and marine sources (1). Marine OM is generally more nitrogen rich than terrestrial OM. It contains carbohydrates and proteins derived largely from water column organisms, compared with carbohydrates, such as cellulose and lignin, which are derived from vascular plants in the terrestrial component (1, 2). The contributions of these distinct organic pools to marine sediment varies between locations, climates, and geologic times (see, e.g., references 2–5). How these sources impact sedimentary carbon cycling and resident organisms is an area of active research.
The marine sedimentary biosphere holds an estimated 5 × 1029 prokaryotic cells (6, 7). Their metabolisms vary between sediments, depending partially on nutrient, electron acceptor, and electron donor availabilities (8–10). Surface microbial communities, temperature, recalcitrance of sediment OM, and depositional conditions also influence the composition and activities of the sedimentary biosphere (11–15). In near-shore environments, such as inland seas and along continental margins, organic loading to the sediment drives the development of the microbial “anaerobic food web” (see references 16 and 17 and references therein). In organic-rich sediments, microbes in the upper few meters of sediment below the seafloor use electron acceptors [e.g., O2, NO3−, Mn(IV), Fe(III), and SO42−] in order of declining energy yields from organic matter respiration, ending with methanogenesis as the dominant process (18). Throughout the sediment column, heterotrophic metabolisms are critical to breaking down the complex macromolecules and producing smaller organic compounds, which feed into respiration and methanogenesis (19). However, it is difficult to distinguish between heterotrophic pathways in situ due to the large range of bioavailable organic substrates, the micromolar concentrations of substrates and products, the diversity of active microbial populations, and the number of pathways involved in OM remineralization in sediments (20–23). Recently, advances in molecular biology (e.g., metagenomics and metatranscriptomics), enzymatic assays, and organic geochemistry and analytical chemistry have allowed for more detailed studies of environmental OM degradation by microorganisms (24–26).
The Baltic Sea is an ideal location to study microbial organic remineralization due to its distinct and well-defined geological history as well as varied OM concentrations (27) (Fig. 1C). The Baltic Sea is a shallow intracontinental sea which receives terrestrial inputs from rivers and runoff and marine inputs from the North Sea via the Skagerrak and Kattegat bodies of water (28). This has created a salinity gradient both laterally into the Baltic Sea and vertically in the water column (29). Regional anoxia is frequent in the deepest basins, and the sediments are rich in OM due to eutrophication and high sedimentation rates (up to 500 cm per 1,000 years) (30). Towards the end of the last glaciation, the melting of the Scandinavian Ice Sheet caused dramatic environmental changes in the Baltic region. Approximately 16,000 years ago, as the basin was still partially covered by the Scandinavian Ice Sheet, the glacial Baltic Ice Lake started to form (31). Between the start of the Holocene 11,700 years before present (BP) to approximately 10,700 BP, a connection of the ice lake to the North Sea caused a brief brackish phase of the basin, the Yoldia Sea (32). This was followed by the low-primary-productivity freshwater Ancylus Lake phase. Sediments deposited in both the Baltic Ice Lake and the Ancylus Lake were organic-poor clays (32). By circa 9,800 BP, a permanent gateway from the Baltic Sea to the North Sea was established, and the entire basin became a brackish-marine sea with high productivity (33), forming the Littorina Sea phase. Sediments deposited in the Littorina Sea and in the modern Baltic Sea were OM rich, highly reducing, and often methanogenic (27, 34). Overall, the contrasts in depositional conditions within the past ∼16,000 years (lacustrine versus marine, organic-rich versus organic-poor, etc.) create natural gradients that may influence the types and pathways of organic remineralization possible in the present-day microbial communities.
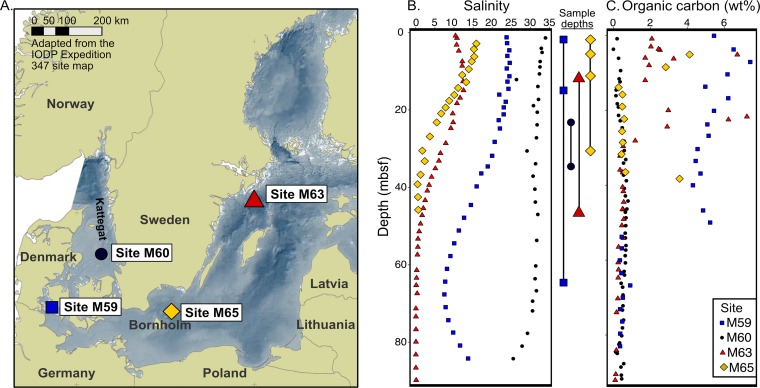
FIG 1.
(A) Map of the samples locations in the Kattegat and Baltic Sea Basin (map adapted from the IODP Expedition 247 site map [2] [coastline: ESRI data and maps, 2005; bathymetry: BALANCE project, www.helcom.fi]). (B and C) Chlorinity-based pore water salinity (B) and total organic carbon (TOC) content (C) in percent dry weight and of sediments from IODP Expedition 347. Sample depths are indicated in the space between panels B and C and correspond to the key in panel C. Values for salinity and TOC were collected and reported as part of IODP Expedition 347 (38).
Previous studies in Baltic Sea sediments have shown that microbial communities vary between sediments deposited during different phases of the Baltic Sea (35). A recent metagenomic analysis of sediments recovered during the Integrated Ocean Drilling Project (IODP) Expedition 347 demonstrated significant differences in the microbial community structure and potential based on deposition, including halogenated compound degradation and C1 metabolisms, such as methane usage and the Wood-Ljungdahl pathway (36). A metatranscriptomics analysis concluded that microbes were active in Holocene-aged sediments up to 42 meters below seafloor (mbsf) (37). The focus of these studies were broad, and neither of these studies examined microbial organic matter mineralization potential in detail.
Here, we investigate sediments from several depths at four sites that differ in organic matter content and depositional histories, as follows: three sites within the Baltic Sea Basin and one site in the Kattegat body, the basin’s marine connection to the North Sea. We present new metagenomic sequencing data from Baltic Sea sediments and reanalyze previously published metagenomes and metatranscriptomes to determine which OM mineralization pathways are present in the sediments, assess which of these pathways are likely active, and connect these heterotrophic metabolisms to sediment facies and geochemistry.
RESULTS
Site description.
Sediment samples were collected from three locations in the Baltic Sea (sites M59, M63, and M65) and one location in the Kattegat region (site M60) (Fig. 1A). The water depths were between 31 m and 437 m, with cored sediment depths from 0.25 m to 67 mbsf (Table 1). Samples taken in the Baltic were deposited either under nonglacial conditions in the Holocene (samples M59E-0.25m, M59E-15m, M63E-12m, M65C-0.25m, M65C-3m, and M65C-10m) or during the Upper Pleistocene or Lower Holocene when the basin was under significant glacial meltwater influence, i.e., during the Baltic Ice Lake phase (samples M59E-67m, M63E-47m, and M65C-30m) (27). The nonglacial samples were deposited during marine phases (the modern Baltic Sea, the Littorina Sea, and the Yoldia Sea) or during the Ancylus Lake phase of the basin (see Fig. S1 in the supplemental material). The samples from M60B in the Kattegat region were marine in origin and were deposited in the Upper Pleistocene after deglaciation and marine transgression of the Kattegat (36).
TABLE 1.
Sample locations and characteristicsa
| Site | Location | Seafloor depth (m) | Below seafloor depth (m) | Depositional conditions | Cl−-based salinity | Total carbon (wt%) | Total organic carbon (wt%) | Concn |
|||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Methane (mM) | Sulfate (mM) | Formate (μM) | Acetate (μM) | Propionate (μM) | Butyrate (μM) | ||||||||
| 59E Little Belt | 55°0.285′N, 10°6.499′E | 37.1 | 0.25 | Holocene marine | 23.30 | 6.40 | 5.40 | 0.25 | 1.61 | 3.95 | 0.96 | 0.17 | |
| 15 | Holocene marine | 24.23 | 5.95 | 4.97 | 1.13 | 0.07 | 10.38 | 20.54 | 4 | 0.43 | |||
| 67 | Glacial lacustrine | 7.48 | 2.17 | 0.91 | 1.76 | 0.01 | 0.74 | 24.04 | 6.61 | 1.04 | |||
| 60B Kattegat | 56°37.204′N, 11°40.229′E | 31.2 | 24 | Marginal marine | 31.72 | 2.08 | 0.48 | 0.00 | 6.59 | 2.73 | 11.85 | 4.19 | 0.42 |
| 37 | Marginal marine | 30.67 | 3.21 | 0.55 | 0.00 | 14.45 | 2.73 | 11.85 | 4.19 | 0.42 | |||
| 63E Landsort Deep | 58°37.330′N, 18°15.240′E | 437.1 | 11 | Holocene marine | 12.03 | 1.76 | 1.53 | 2.33 | 0.01 | 0.00 | 37.38 | 3.03 | 0.2 |
| 47 | Glacial lacustrine | 1.67 | 0.70 | 0.55 | 9.40 | 0.02 | 0.38 | 22.73 | 4.71 | 0.14 | |||
| 65C Børnholm Basin | 55°28.084′N, 15°28.624′E | 84.3 | 0.25 | Holocene marine | 15 | 5 | 4.99 | 0.19 | 2.3 | 2.92 | 1.80 | 1.05 | 0 |
| 3 | Holocene marine | 15.52 | 4.16 | 3.67 | 10.10 | 0.03 | 2.55 | 34.90 | 3.66 | 0 | |||
| 10 | Holocene lacustrine/marine transition | 12.84 | 0.88 | 0.97 | 9.30 | 0.07 | 2.32 | 19.50 | 2.01 | 0.12 | |||
| 30 | Glacial lacustrine | 2.78 | 2.48 | 0.48 | 0.80 | 0.22 | 0.00 | 22.80 | 2.46 | 0.25 | |||
Location data, depositional conditions, salinity, total carbon, total inorganic carbon, alkalinity, methane, and sulfate data were originally published as part of the IODP Expedition 347 post-cruise report and by Andrén et al. (38), except for data for sample M65C-0.25m, which was published by Beulig et al. (39). Formate, acetate, propionate, and butyrate concentrations were measured in this publication, as detailed in Materials and Methods.
Total carbon (TC), total organic carbon (TOC), salinity, and methane were previously reported (27), and data specific to samples discussed herein are summarized here. The TC content of sediment in our samples varied between 0.70% dry weight (d.wt) in sample M63E-47m and 6.40% d.wt in sample M59E-0.25m (Table 1) (27). TOC content ranged from 0.48% d.wt in samples M60B-24m and M65C-30m to 5.40% d.wt in M59E-0.25m (Table 1 and Fig. 1C) (38). Methane was present in all samples except M60B-24m and M60B-37m. The highest measured methane concentration was 10.10 mM, which was in sample M65C-3m (Table 1) (27). Because the cores experienced substantial degassing during sampling, the reported values are minimum in situ methane concentrations (27, 34). Methane in these sediments is biogenic in origin (34, 39). Our Baltic samples (i.e., sites M59, M63, and M65) originated from within or below the main sulfate reduction zone, which is within the top meter of sediments, and the measured sulfate concentrations were therefore generally low (0.01 to 0.22 mM) (39) (Table 1). Samples from site M60 were the exception, with contained sulfate concentrations of 6.59 mM at 24 m and 14.45 at 37 m (Table 1) and no detectable methane until 92.88 mbsf (38). Generally, greater TOC and methane concentrations were observed in the nonglacial samples with high pore water salinities than in samples deposited under glacial conditions or during the Late Pleistocene. Oxygen was not measured in these samples, but in situ chemistry and previous studies indicate that sediments taken in the Baltic and Kattegat regions at these depths beneath the seafloor were anoxic (40, 41).
Pore water formate, acetate, propionate, and butyrate levels were measured (Table 1). Acetate was the most concentrated volatile fatty acid (VFA) measured, with concentrations ranging from 1.38 to 37.38 μM. Propionate concentrations ranged from 0.96 to 6.61 μM. Formate concentrations ranged from below the detection limit (BDL) to 10.38 μM. Butyrate was the least concentrated VFA, with concentrations ranging from BDL to 1.04 μM.
Sequencing and assembly.
Coassembly of metagenomes presented here and metagenomes from a study by Marshall et al. (36) produced 557,851 contigs ≥1,000 bp in length. The coassembly contained a total of 1.07 Gb, with a maximum contig length of 159,111 bp and an average length of 1,923 bp (Table S1). Between 15.43% and 35.95% of metagenomic and metatranscriptomic reads in each sample mapped to the assembly. From the coassembly, 1,477,923 open reading frames (ORFs) were predicted (Table S1). Compared to all protein-coding genes in the InterProScan version 66.0, Pfam version 31.0, and TIGRFAM version 15.0 databases (accessed October 2017), based on sequence similarity, 1,074,069 ORFs were predicted to encode proteins within functional families or putative/hypothetical families in at least one of these databases. The remaining ORFs did not correspond to known or hypothetical genes in these databases.
Protein utilization.
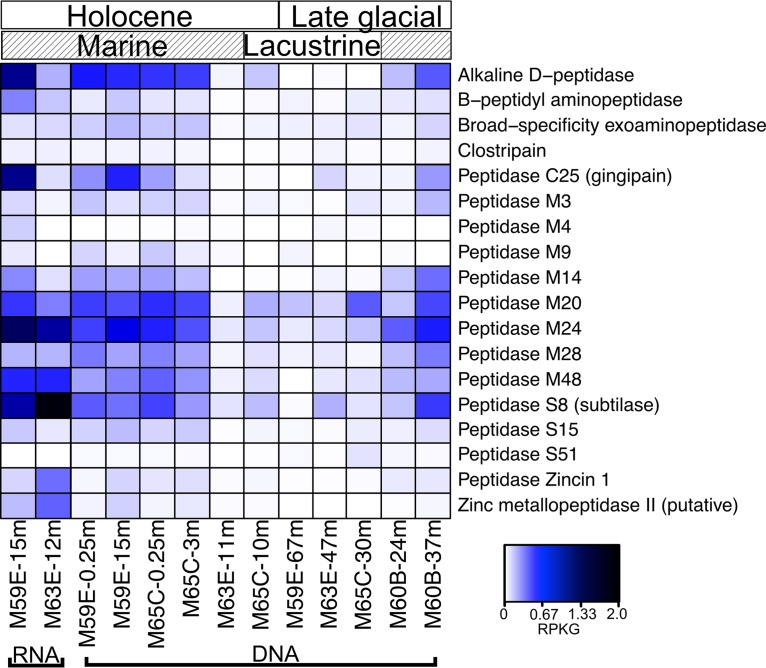
Assembled ORFs were interrogated for sequences that were annotated as peptidase-encoding genes and contained an export signal peptide. ORFs predicted to code for peptidase families M24 (methionine aminopeptidase), S8 (subtilase), and M20 (glutamate carboxypeptidase) were the most abundant putative peptidase-encoding ORFs in total, with ORFs predicted to encode families C25 (gingipain) and M48 (Ste24 endopeptidase), which were also abundant across all samples (Fig. 2). Abundances of ORFs predicted to encode three of these peptidase families (glutamate carboxypeptidase, gingipain, and Ste24 endopeptidase) were significantly positively correlated with TOC content (linear regression, P < 0.05), and methionine aminopeptidase was significantly related to TOC content, salinity, and marine versus lacustrine depositional conditions (Fig. S2). The abundances of ORFs predicted to encode subtilase were not significantly associated with salinity or TOC content but were higher under marine than lacustrine depositional conditions (t test, P = 0.049), as were other less abundant putatively peptidase-encoding ORFs (Fig. S2). All metagenomic reads mapping to the predicted extracellular peptidase-encoding ORFs were significantly more abundant in samples with the greatest TC content (permutational multivariate analysis of variance [PERMANOVA], P = 0.007; Table S3). The relative abundances of these ORFs were also significantly positively correlated with salinity (P = 0.023), marine versus lacustrine depositional conditions salinity (P = 0.033), TOC content (P = 0.039), and formate concentrations (P = 0.021) (Table S3). The transcribed peptidase-encoding ORFs included those for alkaline d-peptidase, peptidase M24, gingipain, and subtilase (Fig. 2).
FIG 2.
Abundances of ORFs which putatively encode extracellular peptidases (listed by MEROPS or Pfam nomenclature) in metagenomes and metatranscriptomes with heatmap color corresponding to RPKG. The type of peptidase putatively encoded in the metagenomes/metatranscriptomes is listed along the y axis. The x axis is arranged by sample type (RNA or DNA) and by the depositional times and environments. Along the top of the heatmap is the time period in which the samples were deposited (Holocene versus late glacial) and the state of the Baltic Sea or Kattegat (marine-influenced versus lacustrine). Marine-influenced samples were deposited during the Yoldia Sea or Baltic Sea phases of the basin or were deposited in the Kattegat body of water, which connects the North Sea and the Baltic Sea. Lacustrine-influenced samples were deposited during the Baltic Ice Lake or Ancylus Lake phases of the basin, when there was no significant influx of seawater to the basin.
Phylum-level (or class level in the case of Proteobacteria) taxonomic assignment of the putative exported peptidase-encoding ORFs revealed sequences that were most similar to multiple bacterial and archaeal lineages (Fig. S3). These lineages included candidate phyla, such as “Candidatus Zixibacteria” and “Candidatus Omnitrophica,” as well as Bathyarchaeota, Calditrichaeota, Planctomycetes, Alphaproteobacteria, and Deltaproteobacteria. Mapping to these ORFs represented up to 16.8% of the total peptidase-encoding ORF abundance (Fig. S3). Most putatively exported peptidase-encoding ORFs were not confidently assigned taxonomy, including the ORFs with mapped transcripts.
Carbohydrate utilization.
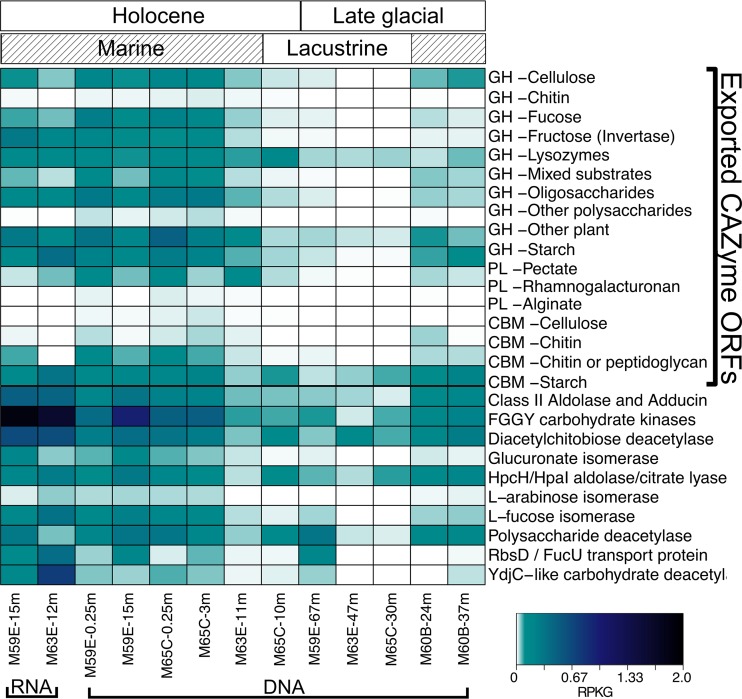
ORFs annotated as coding for proteins that potentially mediate complex carbohydrate degradation (such as ORFs annotated as carbohydrate-active enzymes [CAZymes]) and that contained cellular export signals were examined. Glycoside hydrolases (GHs) are critical proteins in hydrolyzing complex carbohydrates (25). ORFs annotated as GH families with fucosidase, amylase, lysozyme, chitinase, cellulase, and xylanase activities were found (Fig. 3). Collectively, these GHs can degrade carbohydrates from various sources, including plants and algae (42). The most abundant ORFs potentially encoding exported GH families included families 5 (cellulase), 10 (xylanase), 23 to 25 (lysozymes), and 29 (fucosidase) (Fig. 3). The abundances of all of these ORFs annotated as encoding exported GHs except the lysozymes showed significant positive relationships with TOC content (linear modeling, P < 0.05) (Fig. S4). Furthermore, the abundances of putative exported CAZyme-encoding ORFs associated with plant matter, chitin, and starch degradation were significantly associated with marine versus lacustrine depositional conditions (Fig. S5). The abundances of ORFs annotated as encoding lysozymes were significantly correlated with marine Holocene versus glacially influenced depositional conditions (t test, P = 0.00015) (Fig. S4).
FIG 3.
Abundances of ORFs which putatively encode carbohydrate degrading enzymes in metagenomes and metatranscriptomes with heatmap color corresponding to RPKG. The exported CAZyme ORFs bracket on the right side of the heatmap denotes putatively encoded CAZymes (glycoside hydrolases [GHs], polysaccharide lyases [PLs], and carbohydrate binding modules [CBMs]) with export signals and the target substrate(s). The organization of the samples follows Fig. 2.
ORFs annotated as encoding exported carbohydrate binding modules (CBMs), which are associated with GHs and bind target substrates (43), were also found in this study. The most abundant ORFs annotated as encoding CBM found were mostly within families binding cellulose (family 10), chitin (family 5/12), and cell wall material, such as peptidoglycan (family 50) (44–46). Linear modeling showed that chitin-targeting CBM-encoding ORFs were significantly correlated with TOC content (P = 0.0002; Fig. S4). Other putatively exported CAZyme-encoding ORFs found included pectate lyases and alginate lyases, though alginate lyases were in low relative abundance (Fig. 3).
Permutation multivariate analysis of variance tests showed that total exported CAZyme-encoding ORF abundances corresponded most strongly with TOC (P = 0.009) (Table S3). These putative CAZyme-encoding ORF abundances showed weaker but significant correlations to salinity (P = 0.02), marine versus lacustrine depositional environment (P = 0.023), and approximate age of the sediments (P = 0.01) (Table S3).
The two metatranscriptomes showed that ORFs predicted to encode both GHs and CBMs were transcribed in situ (Fig. 3). Transcripts mapping to ORFs predicted to encode cellulases, fucosidases, invertases, lysozymes, plant matter (other than cellulose)-targeting GHs, and oligosaccharide-targeting GHs were relatively abundant, which was similar to the most abundant GH-encoding ORFs in the metagenomes. Of the transcribed CBM ORFs, chitin and peptidoglycan-targeting CBM ORFs were the relatively most abundant, with some transcripts mapping also to starch and plant matter-binding CBM-encoding ORFs (Fig. 3).
Putatively exported CAZyme ORFs were taxonomically assigned to many of the same lineages as the peptidases, such as the Calditrichaeota, “Candidatus Zixibacteria,” Alphaproteobacteria, Planctomycetes, and Chloroflexi (Fig. S3). Additionally, some CAZymes were assigned within the “Candidatus Lokiarchaeota,” and in one sample, M65C-0.25m, reads mapped to ORFs assigned within the bacterial candidate phylum “Candidatus Omnitrophica.” As with the peptidase ORFs, many ORFs were not able to be confidently assigned a taxonomy. The abundance of ORFs assigned within a phylum represented between 0 and 16.1% of the total putative exported CAZyme ORF abundance (Fig. S3). Transcripts mapped to ORFs assigned within Chloroflexi, “Candidatus Lokiarchaeota,” “Candidatus Zixibacteria,” Deltaproteobacteria, and Planctomycetes in M59E-15m and within Chloroflexi and Planctomycetes in M63E-12m (Fig. S3).
Microorganisms can hydrolyze complex carbohydrates into smaller molecules, which are then further metabolized intracellularly (16, 47). ORFs annotated as genes related to these processes were found in all samples (Fig. 3). In M59E-0.25m, M59E-15m, M59E-67m, M63E-11m, M65C-0.25m, and M65C-3m, most relatively abundant ORFs were assigned to the FGGY family, which is a broad family of carbohydrate kinases, such as gluconokinase, xylulokinase, fuculokinase, ribulokinase, and rhamnulokinase (48). ORFs annotated as transporter genes for fucose, which is a sugar subunit of the brown seaweed-produced polysaccharide fucoidan (49), were found in nonglacial metagenomes M59E-0.25m, M59E-15m, M63E-11m, M65C-0.25m, M65C-3m, and M65C-10m and were expressed in the metatranscriptomes but were absent in the glacial samples (Fig. 3). ORFs annotated as containing fucose isomerase genes showed a pattern similar to that of the fucose transporter genes (Fig. 3). ORFs annotated as encoding deacetylases were also found in all metagenomes and expressed in the metatranscriptomes. These included ORFs annotated as encoding diacetylchitobiose deacetylase, which contributes to chitin degradation by removing acetyl groups from diacetylchitobiose (50).
Fermentation.
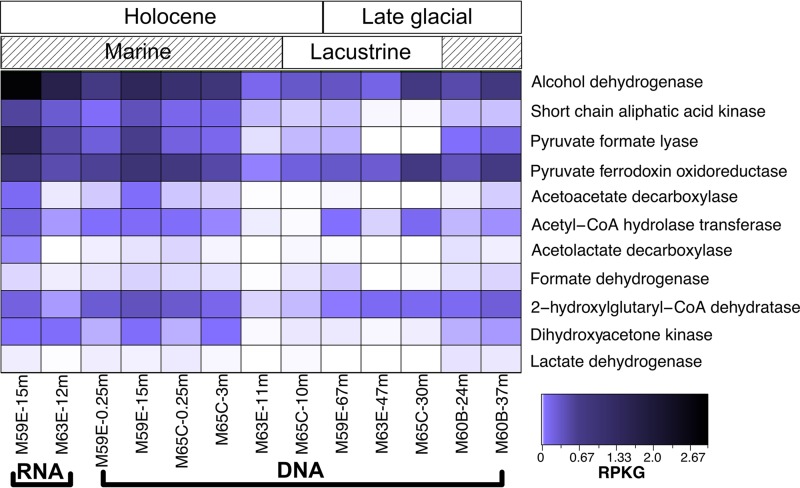
The metagenomes and metatranscriptomes were analyzed for ORFs encoding proteins putatively involved in fermentation. ORFs encoding proteins involved in pyruvate conversion to acetate were abundant, including both pyruvate ferredoxin oxidoreductase and pyruvate formate lyase, and these ORFs were mapped to by the two metatranscriptomic samples (Fig. 4). ORFs putatively encoding pyruvate ferredoxin oxidoreductase were more relatively abundant than ORFs putatively encoding pyruvate formate lyase in all metagenomes (t test, P = 0.0016) (Fig. 4). Putative pyruvate ferredoxin oxidoreductase-encoding ORF abundance was significantly correlated with TC content (linear modeling, P = 0.02) but not TOC content, and putative pyruvate formate lyase-encoding ORFs were significantly positively correlated with TOC content (P = 0.0035) and formate concentration (P = 0.00003) (Fig. S6). In the metatranscriptomes, these two genes were similarly abundant, with ORFs encoding pyruvate formate lyase slightly more abundant in each sample (Fig. 4). ORFs annotated as encoding acetyl-coenzyme A (acetyl-CoA) hydrolase/transferase, which facilitates the production of acetate from acetyl-CoA or the production of acetyl-CoA from acetate and acyl-CoA, and acetate kinases were found in all samples. Acetate kinase-encoding ORF abundances were significantly correlated with TOC content (linear modeling, P = 0.00025), formate concentrations (P = 0.00012) (Fig. S6e and f), and salinity (P = 0.00016).
FIG 4.
Abundances of ORFs which putatively encode fermentation-mediating enzymes in the metagenomes and metatranscriptomes with heatmap color corresponding to RPKG. The organization of the samples follows Fig. 2.
ORFs annotated as coding for alcohol dehydrogenases, which are reversible enzymes that can produce alcohols, such as ethanol, during fermentation (51, 52), were also present and abundant in all samples (Fig. 4). In nonglacial samples, the relative abundance of ORFs annotated as encoding alcohol dehydrogenases was greater than the relative abundance of ORFs encoding pyruvate targeting proteins (Fig. 4). Putative alcohol dehydrogenase ORF abundances were significantly correlated with TC content (linear modeling, P = 0.0022; Fig. S6).
Taxonomic assignments of fermentation ORFs were within diverse phyla, including the Chloroflexi, Planctomycetes, Alphaproteobacteria, Deltaproteobacteria, “Candidatus Omnitrophica,” “Candidatus Lokiarchaeota,” Calditrichaeota, and Bathyarchaeota (Fig. S3). These ORFs represented 0 to 35.2% of the total reads mapped for fermentation genes. Transcripts mapped to fermentation ORFs assigned within Chloroflexi, “Candidatus Lokiarchaeota,” and Calditrichaeota from both metatranscriptomes. Additionally, M59E-15m transcripts mapped to ORFs assigned within Bathyarchaeota, and M63E-12m transcripts mapped to ORFs assigned within Planctomycetes (Fig. S3).
Other present and expressed putatively fermentation-related ORFs included 2-hydroxylglutaryl-CoA dehydratase-encoding ORFs (Fig. 4). 2-Hydroxylgluatryl-CoA dehydratase catalyzes a key step in glutamate fermentation (53, 54). ORFs potentially encoding proteins that oxidize glycerol, dihydroxyacetone kinase, and glycerol dehydratase (55) were found in low abundances in the metagenomes, and at least one of these putative ORF types was expressed in each metatranscriptome. ORFs putatively encoding acetoacetate decarboxylase, which produces acetone and CO2 from acetoacetate during fermentation (56), were present in all Holocene metagenomes (Fig. 4), and their abundances were strongly correlated with TOC content (P = 0.0042). The overall abundance of putative fermentation ORFs in samples was significantly related to TOC content (PERMANOVA, P < 0.05) but not to salinity, marine versus lacustrine sediment deposition, any fatty acid examined here, or sediment age (Table S3), and it generally did not show strong grouping by sample type with clustering analyses (Fig. S7c).
DISCUSSION
Organic matter degradation.
Protein-derived compounds account for >20% of OM in some surface sediments (57, 58). In organic-rich sediments, the microbial genetic potential to degrade macromolecules has previously been demonstrated through genomic analyses (26, 59, 60). In our study, putative peptidase-encoding ORFs with export signal sequences were abundant in marine samples (Fig. 2). The relative abundance of ORFs putatively encoding peptidases was positively correlated with sediment characteristics, including TOC content, salinity, and marine versus lacustrine depositional conditions (Table S3). Recently, Schmidt and Steen incubated Baltic Sea sediment with labeled peptidase substrates and showed that extracellular peptidases from site M59 were active down to 55 mbsf in the organic-rich Holocene sediment (61). Without further information about the origin of the ORFs, such as metagenomic binning or single-cell genome amplification, taxonomic assignments of ORFs are solely based on similarity to previously characterized gene sequences. However, the suggested taxonomies of some of the putative extracellular peptidase-encoding ORFs are similar to those found previously in marine sediments. For example, some peptidase-encoding ORFs here were assigned within the Bathyarchaeota or Calditrichaeota phyla. Archaeal genomes isolated from Bathyarchaeota from Aarhus Bay at the entrance to the Baltic Sea contained peptidases, including gingipain and clostripain, which also appeared to be active in enzyme assays of whole sediment (62). Marshall et al. (63) showed that members of Calditrichaeota, a bacterial phylum found in the Baltic Sea and in marine sediments globally, also likely degrade extracellular proteins for energy (63). These studies support our findings that both bacterial and archaeal lineages commonly found in marine sediments could mediate peptide degradation in Baltic Sea sediments here.
Our findings are also similar to what was observed in metatranscriptomes from Peru Margin sediment, where transcripts predicted to be extracellular peptidases were abundant and taxonomically assigned to archaeal lineages, including Bathyarchaeota (59). While it is not possible to determine the in situ metabolic capabilities or activities of the microbial communities based on gene predictions alone, the results here and previous findings support the idea that protein degradation for energy acquisition is an important heterotrophic strategy in the marine deep biosphere and, based on metatranscriptomics, was likely active in some of these sediments.
Globally, near-shore environments receive 0.4 pg year−1 of riverine OM, including structural parts of vascular plants composed mainly from lignin, cellulose, and xylan (64). Our analyses here indicate that ORFs predicted to encode CAZymes associated with plant matter degradation were abundant in Baltic Sea sediments, including multiple glycoside hydrolases and carbohydrate binding modules (Fig. 3). The abundances of these genes were positively correlated with TOC content (Fig. S4 and Table S3), though it is unknown what proportion of TOC is bioavailable to microbial activity or how it varies between sediment samples. Many CAZyme genes were transcribed in Baltic sediments at 12 and 15 mbsf, suggesting active plant matter degradation (Fig. 3). CAZyme-encoding genes have been found in estuarine sediments (60, 65), in deep-sea sediments (66), and in the top 10 cm of sediment from the Landsort Deep (Baltic Sea) (67). Recently, genes encoding proteins that mediate plant-derived OM degradation were found transcribed in Peru Margin sediments (59). Our results demonstrate a genetic potential for plant matter degradation deep into Baltic sediments and that these genes are actively transcribed to depths of at least 15 mbsf.
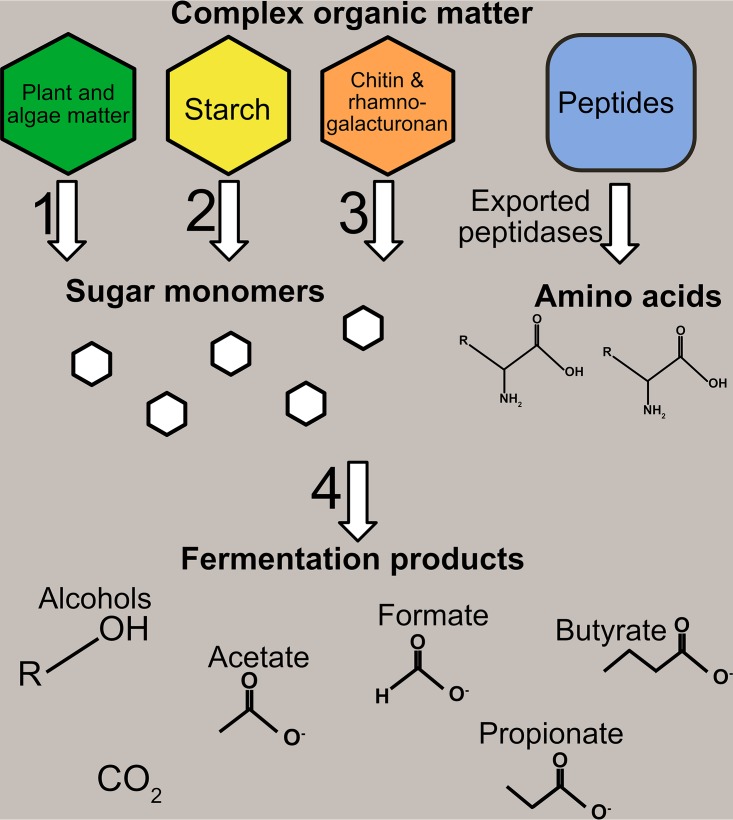
Macroalgae are primary producers which can rapidly produce biomass, which is then deposited in coastal sediments (up to 3 kg C m−2 year−1) (68). In Holocene sediments examined here, putative alginate lyase-encoding ORFs were found in low but detectable numbers at sites M59E, M63E, and M65C and were expressed in M63E-12mbsf and 59E-15mbsf. However, they were not found in sites associated with glacial conditions, which were deposited before the major postglacial establishment of brown algae in the Baltic (69). Brown algae are typically marine seaweeds which produce fucoidan (70). ORFs putatively encoding fucosidases were abundant in sediments deposited during marine-brackish periods, and these ORFs were expressed in the metatranscriptomes. Similarly, we found that ORFs putatively encoding chitinase, which breaks down chitin (71–73), and chitin-binding CBMs were more abundant in samples deposited during periods of marine inputs. Chitin is a major component of the exoskeleton of arthropods and is abundant in marine systems (74). It short, it appears that diverse microbial potential for OM degradation is present in Baltic Sea sediments (Fig. 5). However, due to uncertainties in omics-based studies, experimental evidence is needed to verify that these communities are able to carry out these suggested activities.
FIG 5.
Schematic overview of OM degradation pathways investigated in this study. Assignments of potential ORFs involved in the arrow numbers: 1, fucose, cellulose, and other plant matter GHs, pectate and alginate polysaccharide lyases, cellulose and starch CBMs, glucodextranase, FGGY carbohydrate kinase, l-arabinose isomerase, l-fucose isomerases, pectinesterase, glucuronate isomerase, aldolases, fucose transporters, and polysaccharide deacetylase; 2, starch GHs, starch CBMs, and alpha-amylase/4-alpha-glucanotransferase; 3, chitin GHs, rhamnogalacturonan PLs, chitin CBMs, YdjC-like carbohydrate deacetylase, and diacetylchitobiose deacetylase; 4, ORF assignments from Fig. 4.
Fermentation potential.
ORFs putatively related to fermentative metabolism were ubiquitous in all samples here, were likely taxonomically diverse (Fig. S3), and included those mediating anaerobic fermentation of various substrates (Fig. 4). Particularly abundant were ORFs which putatively confer the ability to break down pyruvate into acetate, i.e., pyruvate ferredoxin oxidoreductase and pyruvate formate lyase. In addition to being among the most abundant genes in the metagenomes, they were also present in the two metatranscriptomes (Fig. 4). These observations agreed with our findings that acetate concentrations in the sediments were higher than those of the other VFAs measured here (Table 1), though neither of these ORF abundances significantly correlated with acetate concentrations. This could be due to the control of VFA turnover by consumers as opposed to producers of VFAs, i.e., fermenters (75).
Alcohol dehydrogenase-encoding ORFs were similarly abundant as ORFs putatively encoding acetate-producing proteins in the metagenomes and were more abundant in the two metatranscriptomes (Fig. 4). In bacteria, archaea, and the yeast Saccaromyces cerevisiae, alcohol dehydrogenase is usually a reversible enzyme, which can produce or break down various alcohols, such as ethanol (51, 52, 76–78). Though we were unable to determine if the dominant function here was alcohol production or consumption, these results indicated that alcohol turnover was an important aspect of carbon cycling in the sedimentary subsurface. Sediments from other marine environments, including methane-rich cold seeps, also have an abundance of alcohol-producing genes (79). Direct measurements within the top meter of Gulf of Mexico sediments revealed methanol and ethanol concentrations up to 69 μM and 43 μM, respectively (80). Furthermore, laboratory studies of fermentation occurring in sediments collected from tidal flats found that ethanol was the second most concentrated fermentation product (acetate was the most concentrated) for at least the first week of the experiment (81). In light of these findings and the recent development of a method to detect micromolar amounts of methanol and ethanol from marine sediments (80), future studies of sediment alcohol dynamics are warranted.
Impacts of depositional condition on microbial community function.
The deglaciation of the Baltic Sea Basin during the late Pleistocene and fluctuating water column conditions throughout the Holocene are reflected in the lithology and in situ geochemistry of Baltic Sea sediment (38). We observed major differences between marine and lacustrine, and organic-rich and organic-poor sediments in the types of possible carbon catabolism, including carbohydrate degradation and extracellular protein degradation. The abundances of these OM degradation ORFs generally were higher in the high organic nonglacial sediments than in the glacial samples (Fig. 2 and 3). However, gene abundances at site M60B, most notably within the putative CAZyme- and peptidase-encoding ORFs, often did not follow the correlation between TOC content and ORF abundances (Fig. 2, 3, and S7a and b). Site M60B is located outside the Baltic Sea Basin in the Kattegat region, and the samples examined here from 24 and 37 mbsf were deposited during the Kattegat deglaciation ca. 15,900 to 16,500 years BP (82). During this period, the Kattegat water column was marine-brackish due to inputs from the North Sea and glacial meltwaters (82), whereas the Baltic Sea Basin was cut off from marine inputs.
One explanation for the abundances of putative CAZyme- and peptidase-encoding ORFs in site 60B could be the depositional history. Most of the marine-influenced samples were from the Holocene (M59E-0.25m, M59E-15m, M65C-0.25m, M65C-3m, and M63E-11m), but the marine-influenced M60B samples were deposited during the Late Pleistocene as the Scandinavian Ice Sheet was retreating (27). During the Late Pleistocene, site M60 received inputs from both glacial meltwater runoff and from the North Sea, and microfossil analyses indicate significant deposition of terrestrial material from glacial meltwater, including Cretaceous-age microfossils (38). Pollen analyses have shown that the easily degradable pollen was underrepresented in the M60B samples relative to pollen that is more resistant to degradation. This was interpreted as extensive degradation of the pollen in situ. This suggests that OM mineralization was active in these sediments postdeposition, and geochemistry indicates that some microbial activity is still occurring in situ (38). In contrast, the other glacially influenced samples were deposited during times with low-water-column productivity and show little evidence of current in situ OM remineralization, as indicated by in situ geochemistry (Table 1). In short, it appears that the depositional conditions influenced modern geochemistry, such as the amount of carbon in sediments, which influences present-day abundances of microbial community genetic potential to break down macromolecules for metabolic use.
The finding that depositional conditions influence microbial community organic matter degradation potential highlights previous observations not only in Baltic Sea sediments (35, 36), but in Arabian Sea sediments as well (83). Sediments in the Arabian Sea, like in the Baltic Sea, exhibit an order of magnitude variation in TOC content corresponding to climate fluctuations over a glacial-interglacial cycle (83). Metagenomic analysis revealed that microbial protein degradation genetic potential was significantly correlated with TOC content in the interglacial sediments in Arabian Sea sediments (83). Similarly, lacustrine sediments from Laguna Potrok Aike, Argentina, were deposited under varied salinity and water column productivity during the last glacial-interglacial cycle (84). Based on 16S rRNA gene profiling, sediment microbial communities were found to vary significantly with these parameters, and it was concluded that climate-related depositional conditions played a role in shaping the subsurface microbial community (84). The culmination of these studies and others (85) with the current study here indicates that depositional conditions and the associated geochemical or lithological conditions impact sediment microbial communities and their metabolic potential postdeposition in a variety of aquatic environments.
In summary, we examined microbial community genetic potential and, in two samples, transcriptional activity to determine the types and relative abundances of microbial heterotrophy in Baltic Sea Basin sediments. We determined that in the organic-rich sediments, there was genetic potential for multiple metabolic strategies, including protein degradation, complex carbohydrate usage, and fermentation (Fig. 5). Furthermore, based on metatranscriptomic analyses, these pathways were active in the two samples. Fermentation potential, including the potential for alcohol production, was ubiquitous in all sediments examined. Finally, the abundance patterns of carbohydrate-active-enzyme- and peptidase-encoding genes indicated that both in situ and sediment depositional conditions are important in determining the types of organic mineralization potential present.
MATERIALS AND METHODS
Sample collection.
Samples M59E-15m, M59E-67m, M60B-24m, M60B-37m, M63E-11m, M63E-47m, M65C-3m, M65C-10m, and M65C-30m were collected by the Integrated Ocean Drilling Program (IODP) aboard the MSP Greatship Manisha in September to November 2013. The sediment cores were taken by advanced piston coring. On board, cores were cut into 1.5-m sections, sampled for perfluorinated chemical (PFC) contamination (to assess the amount of drilling disturbance), scanned with a fast-track multiple-scanning core logger, and sectioned into whole round cores in a 12°C microbiology container onboard the ship. Sediment cores for nucleic acid analyses were immediately frozen at −80°C on ship and shipped to land-based laboratories on dry ice (cf. reference 38).
To sample the top few meters of sediment, which were not recovered in the drilling process, two cruises lead by the Center for Geomicrobiology at Aarhus University on the R/V Aurora were undertaken. In September 2014, site M59 was revisited. Sample M59E-0.25cm was collected through gravity coring, followed by subsampling through windows cut into the sediment core liner and the insertion of sterile 20-ml syringes with the ends cut off. In June, 2016, site M65C was revisited, and sample M65C-0.25m was collected through Rumohr coring (as in reference 39). Subsampling was similar to that with sample M59E-0.25m, in which sterile cut-off 20-ml syringes were used to subsample the 25-cm-depth horizon. Care was taken to avoid potential seawater contamination by visually inspecting the cores for seawater intrusion and extracting DNA from sediment collected from the interior of the core. All samples were immediately frozen at −80°C on ship and were shipped to the United States on dry ice.
Volatile fatty acid analysis.
Pore water samples for volatile fatty acid (VFA) analysis were retrieved with Rhizon soil moisture samplers (Rhizosphere Research Products, Wageningen, The Netherlands) (86) or were obtained by a hydraulic press (87) according to IODP standard protocols if the sediment was to compacted. Rhizon samplers were precleaned with 50 ml Milli-Q water (ultrapure, type 1) and stored in vacuum-sealed gas-tight bags (24). The samples were stored at −80°C in 4-ml borosilicate glass vials (Zinsser Analytic, Germany) that were previously baked for 5 h at 450°C. Prior to the analysis, the samples were defrosted and filtered through disposable Acrodisc 13-mm ion chromatography (IC) syringe filters (pore size, 0.2 μm) that were rinsed with 10 ml Milli-Q (ultrapure, type I) water directly before use. The first 0.5 ml of pore water after filtration was discarded, and a second 0.5 ml was used for analysis. VFA concentrations, including formate, acetate, butyrate, and propionate, were measured by two-dimensional IC mass spectrometry (2D IC-MS), as described in detail by Glombitza et al. (24). Briefly, in this method, the first IC dimension was used to separate inorganic ions, such as chloride, from VFAs. VFAs were trapped on a concentrator column and subsequently separated in the second IC dimension. Quantification was achieved by the mass spectrometer in the single ion monitoring (SIM) mode. The detection limits were 0.37 μM for formate, 0.19 μM for acetate, 0.12 μM for propionate, and 0.09 μM for butyrate. Quantification was achieved by a 3-point calibration with external standards of a mixture of VFAs (formate, acetate, and propionate) at different concentrations (i.e., 200, 500, and 800 μg liter−1) in an International Association for Physical Sciences of the Ocean (IAPSO) seawater standard (Ocean Scientific International Ltd. [OSIL], UK).
Other geochemical data for IODP cores (all samples here except M59E-0.25m and M65C-0.25m) were collected and analyzed as described in reference 38. Briefly, Rhizon soil moisture samplers and core squeezers were used to retrieve sediment pore water. Sulfate and Cl− were measured via ion chromatography using a Metrohm 882 compact ion chromatograph (Herisau, Switzerland) at the University of Bremen. Methane samples were collected from fresh core material, extruded into 8 ml of 1 M NaOH-filled glass vials, shaken and equilibrated, and measured on an A7890 gas chromatograph (Agilent Technologies, Santa Clara, CA, USA) (34, 38). TC and TOC were sampled from 10 cm3 of freeze-dried and ground sediment. TC measurements were derived from approximately 65 mg of sample that was combusted, and evolved CO2 was measured on a CS-300 carbon-sulfur analyzer (Leco Corporation, St. Joseph, MI, USA). TOC was measured from 65 mg of 12.5% HCl decalcified sediment, which was then heated, and evolved CO2 was measured as described above (38).
For samples M59E-0.25cm and M65C-0.25cm, geochemical analyses were performed as described previously (39). Rhizon samplers extracted sediment pore water, which was then acidified and measured at the University of Aarhus (88). Methane samples were collected immediately after core retrieval, transferred into vials with 4 ml of saturated NaCl, capped, stored at −20°C, and measured on an SRI 310C gas chromatograph equipped with an SRI 310C flame ionization detector (SRI Instruments, Torrance, CA, USA) (39).
Extraction.
DNA from samples M59E-0.25m, M59E-15m, M65C-0.25m, M65C-3m, and M65C-10m was extracted from sediment using the DNeasy PowerMax soil kit (Mo Bio Laboratories, Carlsbad, CA). Frozen sediment was chipped from whole round cores (M59E-15m, M65C-3m, and M65C-10m) or cut-off syringes (M59E-0.25m and M65C-0.25m) in a dedicated clean room at Texas A&M University Corpus Christi. All instruments used were treated with ethanol and flame-sterilized, and the edges of the sediment core were avoided during collection. Researchers wore face masks and hair nets to avoid sample contamination. Between 5 and 10 g of sediment was extracted for each sample. The manufacturer’s protocol was followed, including the final concentration step and resuspension of DNA in 100 μl of molecular-biology-grade water. Sample-free negative controls (kit blanks) were processed and sequenced alongside the samples. These negative controls were below the detection limit (0.5 ng DNA μl−1) when measured using the Qubit DS high-sensitivity kit and did not amplify when subject to PCR of the 16S rRNA gene and visualized on a 1% agarose gel.
Sequencing.
Metagenomes from samples M59E-0.25m, M59E-15m, and M65C-0.25m were sequenced at the Marine Biological Laboratories (Woods Hole, MA, USA). Metagenomic library preparation and sequencing followed the Census of Deep Life protocol as described by Vineis et al. (89). The modifications to the Vineis protocol were that the sequencing platform was the NextSeq (Illumina, San Diego, CA, USA), which produced 150-bp-long paired-end reads, and no microbiome enrichment step was conducted. The insert sizes were 170 bp.
Metagenomes from samples M65C-3mbsf and M65C-10mbsf were sequenced at the Research and Testing Labs (RTL; Lubbock, TX, USA). Libraries were prepared using the HyperPlus kit (Kapa Biosystems, Wilmington, MA, USA), according to the manufacturer’s instructions, with the following modifications: ligation was increased to 1 h at room temperature, postligation cleanup used 0.75× beads, and the postamplification cleanup bead concentration was increased to 0.7×. Libraries were sequenced on the HiSeq 2500 system (Illumina, San Diego, CA, USA), producing 150-bp-long paired-end reads. The insert sizes were 170 bp.
See Table S1 for sequencing statistics. Note that the metagenome M63E-12m and the metatranscriptome M63E-12m were both extracted from sediment core M63E-6H2 (IODP nomenclature), which spanned from 10 to 12 mbsf (36–38). To maintain consistency with these prior publications, the names of the metagenome and metatranscriptome from this core used in this publication are M63E-12m and M63E-11m, respectively. The differences in sequencing coverages between the metagenomes presented here and the previously published metagenomes could cause some bias in detection of low-abundance ORFs. Additionally, assembling these samples individually was not ideal due to the low sequencing coverage in the samples from the study by Marshall et al. (36), making coassembly of all metagenomes the optimal choice and consistent with previous studies (90–92).
Bioinformatics. (i) Quality control and assembly.
Reads were trimmed using the program Trim Galore! version 0.4.3 (Babraham Bioinformatics, Cambridge, United Kingdom) in paired-end read mode, with a minimum quality score of 25, a maximum of 4 low-quality bases before the read was trimmed, and the read length must be a minimum of 80 bp long posttrimming. Samples were deduplicated using Super Deduper version 2.0, with default settings (starting location of 10 bp, 25 bp in the unique identification [ID]) (93). All metagenomes were coassembled using MEGAHIT version 1.0.3-29-g707d683, with a minimum contig size of 1,000 bp (94, 95). Default k-mer sizes of 21, 29, 39, 59, 79, 99, 119, and 141 were used for assembly. Contig names were simplified using anvi-script-reformat-fasta in anvi’o v2.4.0 (96).
(ii) Read mapping and profile generation.
Metagenomic and metatranscriptomic reads were mapped to the assembled contigs using Bowtie2 version 2.2.5 using the “sensitive” end-to-end setting (97). The resulting .sam files were converted to bam files using SAMtools version 1.5, and these files were converted to anvi’o-compatible .bam files in anvi’o. An anvi’o database was created from the contigs, which included ORF determination using Prodigal (98). Each sample was profiled against the contig database using the anvi’o command anvi-profile. A full project database was constructed from these profiles and included information about gene coverage and detection (percentage over which the ORF was mapped by reads). Tables with gene coverage by sample and gene detection by sample were exported using the anvi’o command anvi-export-gene-coverage-and-detection. In this command, gene coverage is reported as coverage of each base pair of the gene from mapped reads, divided by the length of the gene, and these values were then normalized to reads per kilobase per billion (RPKG). Only genes with at least 50% detection, referring to at least 50% of the gene being represented by at least 1× coverage prenormalization, were considered (96).
Taxonomic and function assignments of genes.
ORFs were exported as amino acid sequences from the contigs for taxonomic and functional assignment using the anvi’o command anvi-get-aa-sequences-for-gene-calls. Function was assigned to assembled genes using InterProScan version 5.26-65.0 (99) against the Pfam version 31.0 (100) and TIGRFAM version 15.0 databases (101) (accessed September 2017) using the precalculated lookup service. Functional assignments were exported as a tab-separated file and parsed in R. Genes annotated as peptidase or CAZyme coding were screened for export peptides using SignalP targeted for bacteria (Gram positive and negative) and eukaryotes (102), and were screened using PSORTb targeted for archaea (103). Protein families of interest were manually searched in the Pfam database in August to November 2017. Pathway reconstruction was based on pathways in the KEGG (104) and Metacyc databases (105) and published literature. Putative peptidase-encoding gene functions were described using the MEROPS peptidase nomenclature (106). Glycoside hydrolase functions were inferred from the Pfam GH assignment using the functions as described in the CAZYpedia and in published literature (25, 42). These functions were summarized by putative substrate in Fig. 3. For a complete list of genes examined and their Pfam numbers, see Table S2.
ORF sequences were compared to the NCBI nonredundant database (accessed December 2016) using the blastp mode of DIAMOND version 0.8.36 (107) with “sensitive” setting and allowing only one match per sequence allowed. The DIAMOND results were uploaded to MEGAN version 6.10.2, and taxonomy was assigned using the weighted lowest common ancestor (LCA) assignment algorithm with a minimum support percent identity of 0.3 (i.e., a taxon must have at least 0.3%, or 2,888 ORFs, assigned to be considered a “real” hit, as suggested for mixed communities in the user manual) (108, 109). Taxonomic assignments for ORFs were exported as a tab-separated file and parsed in R.
Statistical analyses.
All statistics, including linear modeling, canonical correspondence analyses, Student’s t tests, and permutational analysis of variance (PERMANOVA), were performed using the vegan package version 2.4-4 (110) in R version 3.4.2, and the type of statistical test used is indicated in the text. For tests involving geochemical parameters, such as salinity or TOC content, values listed in Table 1 were used. Sediment age was estimated from previously published scientific literature (111, 112). Marine versus lacustrine conditions were defined by the stage of the Baltic Sea Basin during sediment deposition, i.e., Baltic Ice Lake and Ancylus Lake were lacustrine and the Yoldia Sea, Littorina Sea, and the modern Baltic Sea were marine (Table 1 and Fig. S1). Both Kattegat samples were marine. All P values reported herein were determined to be statistically significant at a value of less than 0.05. The ggplot2 package (113) was used to create graphics.
We recognize that this data set is a relatively small sample set (n = 11), and this may reduce the robustness of statistical analyses. The application of statistical analyses nevertheless has merit even in environments of low sample density and has previously been applied successfully in the marine deep biosphere (see, e.g., references 35 and 61). The variation in sequencing coverage across each sample could bias results by underestimating low-abundance genes in samples sequenced with lesser coverage, and our coassembly could be biased towards the samples that contained a greater abundance of sequences. However, our results consistently indicated that the abundance and type of OM degradation potential found in the metagenomes were related to several sediment characteristics, including TOC content and depositional facies.
Data availability.
Metagenomic data for M59E-0.25m, M59E-15m, M65C-0.25m, M65C-3mbsf, and M65C-10mbsf can be found in the National Center for Biotechnology Information (NCBI) under BioProject number PRJNA433242. Metagenomes from all other samples here were sequenced as described by Marshall et al. (36) and were retrieved from the NCBI SRA project SRP068645. Metatranscriptomes were sequenced as described by Zinke et al. (37) and archived under the NCBI SRA project SRP108285.
Supplementary Material
ACKNOWLEDGMENTS
We thank the entire science party and crew of Integrated Ocean Drilling Program Expedition 347: Baltic Sea Paleoenvironment and the MSP Greatship Manisha, of the Geomicrobiology of the Skagerrak and Kattegat cruise (2014), and of the AUBO16 cruise and R/V Aurora. We greatly appreciate the help of Felix Beulig, André Pellerin, Alex Michaud, Gilad Antler, Susanne Nielsen, and R/V Aurora captain Torben Vang during sampling. We thank Megan Mullis, Rachel Weisend, Morgan Sobol, and Pratixa Savalia for their assistance with laboratory work; Mike Lee, Elaina Graham, and Benjamin Tully for their bioinformatics insight; and Ian P. G. Marshall, Annie Rowe, and Doug LaRowe for helpful discussions.
This research was supported by the National Science Foundation Biological Oceanography grant OCE-1431598 to J.P.A. (with a subaward to K.G.L.), the Sloan Foundation Deep Carbon Observatory’s Census of Deep Life, and Danish Center for Marine Research Grant “Cryptic Biogeochemistry in the Bornholm Basin” to H.R. L.A.Z. was supported by a USSSP Schlanger Fellowship.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.02164-18.
REFERENCES
- 1.Mackenzie FT, Lerman A, Andersson AJ. 2004. Past and present of sediment and carbon biogeochemical cycling models. Biogeosci Discuss 1:27–85. doi: 10.5194/bgd-1-27-2004. [DOI] [Google Scholar]
- 2.Calvert SE. 1987. Oceanographic controls on the accumulation of organic matter in marine sediments. Geol Soc Spec Publ 26:137–151. doi: 10.1144/GSL.SP.1987.026.01.08. [DOI] [Google Scholar]
- 3.Stein R. 1990. Organic carbon content/sedimentation rate relationship and its paleoenvironmental significance for marine sediments. Geo-Marine Lett 10:37–44. doi: 10.1007/BF02431020. [DOI] [Google Scholar]
- 4.Aller RC, Blair NE. 2004. Early diagenetic remineralization of sedimentary organic C in the Gulf of Papua deltaic complex (Papua New Guinea): net loss of terrestrial C and diagenetic fractionation of C isotopes. Geochim Cosmochim Acta 68:1815–1825. doi: 10.1016/j.gca.2003.10.028. [DOI] [Google Scholar]
- 5.Tao S, Eglinton TI, Montluçon DB, McIntyre C, Zhao M. 2016. Diverse origins and pre-depositional histories of organic matter in contemporary Chinese marginal sea sediments. Geochim Cosmochim Acta 191:70–88. doi: 10.1016/j.gca.2016.07.019. [DOI] [Google Scholar]
- 6.Kallmeyer J, Pockalny R, Adhikari RR, Smith DC, D’Hondt S. 2012. Global distribution of microbial abundance and biomass in subseafloor sediment. Proc Natl Acad Sci U S A 109:16213–16216. doi: 10.1073/pnas.1203849109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Parkes RJ, Cragg B, Roussel E, Webster G, Weightman A, Sass H. 2014. A review of prokaryotic populations and processes in sub-seafloor sediments, including biosphere:geosphere interactions. Mar Geol 352:409–425. doi: 10.1016/j.margeo.2014.02.009. [DOI] [Google Scholar]
- 8.Röling WFM, Milner MG, Jones DM, Lee K, Daniel F, Swannell RJP, Head IM. 2002. Robust hydrocarbon degradation and dynamics of bacterial communities during nutrient-enhanced oil spill bioremediation. Appl Environ Microbiol 68:5537–5548. doi: 10.1128/AEM.68.11.5537-5548.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lloyd KG, Lapham L, Teske A. 2006. An anaerobic methane-oxidizing community of ANME-1b archaea in hypersaline Gulf of Mexico sediments. Appl Environ Microbiol 72:7218–7230. doi: 10.1128/AEM.00886-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Flores GE, Campbell JH, Kirshtein JD, Meneghin J, Podar M, Steinberg JI, Seewald JS, Tivey MK, Voytek MA, Yang ZK, Reysenbach A-L. 2011. Microbial community structure of hydrothermal deposits from geochemically different vent fields along the Mid-Atlantic Ridge. Environ Microbiol 13:2158–2171. doi: 10.1111/j.1462-2920.2011.02463.x. [DOI] [PubMed] [Google Scholar]
- 11.Inagaki F, Nunoura T, Nakagawa S, Teske A, Lever M, Lauer A, Suzuki M, Takai K, Delwiche M, Colwell FS, Nealson KH, Horikoshi K, D’Hondt S, Jørgensen BB. 2006. Biogeographical distribution and diversity of microbes in methane hydrate-bearing deep marine sediments on the Pacific Ocean Margin. Proc Natl Acad Sci U S A 103:2815–2820. doi: 10.1073/pnas.0511033103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Finke N, Jorgensen BB. 2008. Response of fermentation and sulfate reduction to experimental temperature changes in temperate and Arctic marine sediments. ISME J 2:815–829. doi: 10.1038/ismej.2008.20. [DOI] [PubMed] [Google Scholar]
- 13.Hamdan LJ, Coffin RB, Sikaroodi M, Greinert J, Treude T, Gillevet PM. 2012. Ocean currents shape the microbiome of Arctic marine sediments. ISME J 7:685–696. doi: 10.1038/ismej.2012.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Canion A, Overholt WA, Kostka JE, Huettel M, Lavik G, Kuypers MMM. 2014. Temperature response of denitrification and anaerobic ammonium oxidation rates and microbial community structure in Arctic fjord sediments. Environ Microbiol 16:3331–3344. doi: 10.1111/1462-2920.12593. [DOI] [PubMed] [Google Scholar]
- 15.Starnawski P, Bataillon T, Ettema TJG, Jochum LM, Schreiber L, Chen X, Lever MA, Polz MF, Jørgensen BB, Schramm A, Kjeldsen KU. 2017. Microbial community assembly and evolution in subseafloor sediment. Proc Natl Acad Sci U S A 114:2940–2945. doi: 10.1073/pnas.1614190114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Middelburg JJ, Vlug T, Jaco F, van der Nat WA. 1993. Organic matter mineralization in marine systems. Glob Planet Change 8:47–58. doi: 10.1016/0921-8181(93)90062-S. [DOI] [Google Scholar]
- 17.Jorgensen BB. 2006. Marine geochemistry, p 169–206. In Schulz HD, Zabel M (ed), Bacteria and marine biogeochemistry, 2nd ed Springer, Berlin, Germany. [Google Scholar]
- 18.Froelich PN, Klinkhammer GP, Bender ML, Luedtke NA, Heath GR, Cullen D, Dauphin P, Hammond D, Hartman B, Maynard V. 1979. Early oxidation of organic matter in pelagic sediments of the eastern equatorial Atlantic: suboxic diagenesis. Geochim Cosmochim Acta 43:1075–1090. doi: 10.1016/0016-7037(79)90095-4. [DOI] [Google Scholar]
- 19.Alperin MJ, Albert DB, Martens CS. 1994. Seasonal variations in production and consumption rates of dissolved organic carbon in an organic-rich coastal sediment. Geochim Cosmochim Acta 58:4909–4930. doi: 10.1016/0016-7037(94)90221-6. [DOI] [Google Scholar]
- 20.Wellsbury P, Parkes RJ. 1995. Acetate bioavailability and turnover in an estuarine sediment. FEMS Microbiol Ecol 17:85–94. doi: 10.1111/j.1574-6941.1995.tb00133.x. [DOI] [Google Scholar]
- 21.Nealson KH. 1997. Sediment bacteria: who’s there, what are they doing, and what’s new? Annu Rev Earth Planet Sci 25:403–434. doi: 10.1146/annurev.earth.25.1.403. [DOI] [PubMed] [Google Scholar]
- 22.Arnosti C, Bell C, Moorhead DL, Sinsabaugh RL, Steen AD, Stromberger M, Wallenstein M, Weintraub MN. 2014. Extracellular enzymes in terrestrial, freshwater, and marine environments: perspectives on system variability and common research needs. Biogeochemistry 117:5–21. doi: 10.1007/s10533-013-9906-5. [DOI] [Google Scholar]
- 23.Arnosti C. 2011. Microbial extracellular enzymes and the marine carbon cycle. Annu Rev Mar Sci 3:401–425. doi: 10.1146/annurev-marine-120709-142731. [DOI] [PubMed] [Google Scholar]
- 24.Glombitza C, Pedersen J, Røy H, Jorgensen BB. 2014. Direct analysis of volatile fatty acids in marine sediment porewater by two-dimensional ion chromatography-mass spectrometry. Limnol Oceanogr Methods 12:455–468. doi: 10.4319/lom.2014.12.455. [DOI] [Google Scholar]
- 25.Berlemont R, Martiny AC. 2016. Glycoside hydrolases across environmental microbial communities. PLoS Comput Biol 12:e1005300. doi: 10.1371/journal.pcbi.1005300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Steen AD, Kevorkian RT, Bird JT, Dombrowski N, Baker BJ, Hagen SM, Mulligan KH, Schmidt JM, Webber AT, Alperin MJ. 2016. Extracellular peptidases in subsurface sediments of the White Oak River estuary, NC, suggest microbial community adaptation to oxidize degraded organic matter. bioRxiv doi: 10.1101/080671. [DOI]
- 27.Andrén T, Barker Jørgensen B, Cotterill C, Green S, the IODP Expedition 347 Scientific Party. 2015. IODP expedition 347: Baltic Sea basin paleoenvironment and biosphere. Sci Dril 20:1–12. doi: 10.5194/sd-20-1-2015. [DOI] [Google Scholar]
- 28.Andersson PS, Wasserburg GJ, Ingri J. 1992. The sources and transport of Sr and Nd isotopes in the Baltic Sea. Earth Planet Sci Lett 113:459–472. doi: 10.1016/0012-821X(92)90124-E. [DOI] [Google Scholar]
- 29.Kullenberg G, Jacobsen TS. 1981. The Baltic Sea: an outline of its physical oceanography. Mar Pollut Bull 12:183–186. doi: 10.1016/0025-326X(81)90168-5. [DOI] [Google Scholar]
- 30.Conley DJ, Björck S, Bonsdorff E, Carstensen J, Destouni G, Gustafsson BG, Hietanen S, Kortekaas M, Kuosa H, Markus Meier HE, Müller-Karulis B, Nordberg K, Norkko A, Nürnberg G, Pitkänen H, Rabalais NN, Rosenberg R, Savchuk OP, Slomp CP, Voss M, Wulff F, Zillén L. 2009. Hypoxia-related processes in the Baltic Sea. Environ Sci Technol 43:3412–3420. doi: 10.1021/es802762a. [DOI] [PubMed] [Google Scholar]
- 31.Houmark-Nielsen M, Kjær KH. 2003. Southwest Scandinavia, 40–15 kyr BP: palaeogeography and environmental change. J Quat Sci 18:769–786. doi: 10.1002/jqs.802. [DOI] [Google Scholar]
- 32.Andrén T, Björck S, Andrén E, Conley D, Zillén L, Anjar J. 2011. The development of the Baltic Sea Basin during the last 130 ka, p 75–97. In Harff J, Björck S, Hoth P (ed), The Baltic Sea Basin. Springer Berlin Heidelberg, Berlin, Germany. [Google Scholar]
- 33.Sternbeck J, Sohlenius G, Hallberg RO. 2000. Sedimentary trace elements as proxies to depositional changes induced by a holocene fresh-brackish water transition. Aquat Geochem 6:325–345. doi: 10.1023/A:1009680714930. [DOI] [Google Scholar]
- 34.Egger M, Hagens M, Sapart CJ, Dijkstra N, van Helmond NAGM, Mogollón JM, Risgaard-Petersen N, van der Veen C, Kasten S, Riedinger N, Böttcher ME, Röckmann T, Jorgensen BB, Slomp CP. 2017. Iron oxide reduction in methane-rich deep Baltic Sea sediments. Geochim Cosmochim Acta 207 IS -:256–276. doi: 10.1016/j.gca.2017.03.019. [DOI] [Google Scholar]
- 35.Lyra C, Sinkko H, Rantanen M, Paulin L, Kotilainen A. 2013. Sediment bacterial communities reflect the history of a sea basin. PLoS One 8:e54326. doi: 10.1371/journal.pone.0054326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Marshall IPG, Karst SM, Nielsen PH, Jorgensen BB. 2017. Metagenomes from deep Baltic Sea sediments reveal how past and present environmental conditions determine microbial community composition. Mar Genomics 37:58–68. [DOI] [PubMed] [Google Scholar]
- 37.Zinke LA, Mullis MM, Bird JT, Marshall IPG, Jorgensen BB, Lloyd KG, Amend JP, Kiel Reese B. 2017. Thriving or surviving? Evaluating active microbial guilds in Baltic Sea sediment. Environ Microbiol Rep 9:528–536. doi: 10.1111/1758-2229.12578. [DOI] [PubMed] [Google Scholar]
- 38.Andrén T, Jorgensen BB, Cotterill C, Green S, Andrén E, Ash J, Bauersachs T, Cragg BA, Fanget AS, Fehr A, Granoszewski W, Groeneveld J, Hardisty D, Herrero-Bervera E, Hyttinen O, Jensen JB, Johnson S, Kenzler K, Kotilainen A, Kotthoff U, Marshall IPG, Martin E, Obrochta S, Passchier S, Quintana Krupinski NB, Riedinger N, Slomp C, Snowball I, Stepanova A, Strano S, Torti A, Warnock J, Xiao N, Zhang R. 2015. Proceedings of the IODP. Integrated Ocean Drilling Program, College Station. [Google Scholar]
- 39.Beulig F, Roy H, Glombitza C, Jørgensen BB. 2018. Control on rate and pathway of anaerobic organic carbon degradation in the seabed. Proc Natl Acad Sci U S A 115:367–372. doi: 10.1073/pnas.1715789115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mort HP, Slomp CP, Gustafsson BG, Andersen TJ. 2010. Phosphorus recycling and burial in Baltic Sea sediments with contrasting redox conditions. Geochim Cosmochim Acta 74:1350–1362. doi: 10.1016/j.gca.2009.11.016. [DOI] [Google Scholar]
- 41.Jorgensen BB, Revsbech NP. 1989. Oxygen uptake, bacterial distribution, and carbon-nitrogen-sulfur cycling in sediments from the Baltic Sea–North Sea transition. Ophelia 31:29–49. doi: 10.1080/00785326.1989.10430849. [DOI] [Google Scholar]
- 42.Lombard V, Golaconda Ramulu H, Drula E, Coutinho PM, Henrissat B. 2013. The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res 42:D490–D495. doi: 10.1093/nar/gkt1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shoseyov O, Shani Z, Levy I. 2006. Carbohydrate binding modules: biochemical properties and novel applications. Microbiol Mol Biol Rev 70:283–295. doi: 10.1128/MMBR.00028-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Notenboom V, Boraston AB, Kilburn DG, Rose DR. 2001. Crystal structures of the family 9 carbohydrate-binding module from Thermotoga maritima xylanase 10A in native and ligand-bound forms. Biochemistry 40:6248–6256. doi: 10.1021/bi0101704. [DOI] [PubMed] [Google Scholar]
- 45.Boraston AB, Bolam DN, Gilbert HJ, Davies GJ. 2004. Carbohydrate-binding modules: fine-tuning polysaccharide recognition. Biochem J 382:769–781. doi: 10.1042/BJ20040892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Martin M, Štefan J. 2008. Domain evolution in the GH13 pullulanase subfamily with focus on the carbohydrate-binding module family 48. Biologia 63:1057. [Google Scholar]
- 47.Arndt S, Jørgensen BB, LaRowe DE, Middelburg JJ, Pancost RD, Regnier P. 2013. Quantifying the degradation of organic matter in marine sediments: a review and synthesis. Earth Sci Rev 123:53–86. doi: 10.1016/j.earscirev.2013.02.008. [DOI] [Google Scholar]
- 48.Zhang Y, Zagnitko O, Rodionova I, Osterman A, Godzik A. 2011. The FGGY carbohydrate kinase family: insights into the evolution of functional specificities. PLoS Comput Biol 7:e1002318. doi: 10.1371/journal.pcbi.1002318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ale MT, Meyer AS. 2013. Fucoidans from brown seaweeds: an update on structures, extraction techniques and use of enzymes as tools for structural elucidation. RSC Adv 3:8131–8141. doi: 10.1039/C3RA23373A. [DOI] [Google Scholar]
- 50.Tanaka T, Fukui T, Fujiwara S, Atomi H, Imanaka T. 2004. Concerted action of diacetylchitobiose deacetylase and exo-β-d-glucosaminidase in a novel chitinolytic pathway in the hyperthermophilic archaeon Thermococcus kodakaraensis KOD1. J Biol Chem 279:30021–30027. doi: 10.1074/jbc.M314187200. [DOI] [PubMed] [Google Scholar]
- 51.Ben-Bassat A, Lamed R, Zeikus JG. 1981. Ethanol production by thermophilic bacteria: metabolic control of end product formation in Thermoanaerobium brockii. J Bacteriol 146:192–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Liang J-J, Zhang M-L, Ding M, Mai Z-M, Wu S-X, Du Y, Feng J-X. 2014. Alcohol dehydrogenases from Kluyveromyces marxianus: heterologous expression in Escherichia coli and biochemical characterization. BMC Biotechnol 14:45. doi: 10.1186/1472-6750-14-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hans M, Sievers J, Müller U, Bill E, Vorholt JA, Linder D, Buckel W. 1999. 2-Hydroxyglutaryl-CoA dehydratase from Clostridium symbiosum. FEBS J 265:404–414. doi: 10.1046/j.1432-1327.1999.00748.x. [DOI] [PubMed] [Google Scholar]
- 54.Schweiger G, Dutscho R, Buckel W. 1987. Purification of 2-hydroxyglutaryl-CoA dehydratase from Acidaminococcus fermentans. Eur J Biochem 169:441–448. doi: 10.1111/j.1432-1033.1987.tb13631.x. [DOI] [PubMed] [Google Scholar]
- 55.Daniel R, Stuertz K, Gottschalk G. 1995. Biochemical and molecular characterization of the oxidative branch of glycerol utilization by Citrobacter freundii. J Bacteriol 177:4392–4401. doi: 10.1128/jb.177.15.4392-4401.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Amador-Noguez D, Brasg IA, Feng X-J, Roquet N, Rabinowitz JD. 2011. Metabolome remodeling during the acidogenic-solventogenic transition in Clostridium acetobutylicum. Appl Environ Microbiol 77:7984–7997. doi: 10.1128/AEM.05374-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wakeham SG, Lee C, Hedges JI, Hernes PJ, Peterson MJ. 1997. Molecular indicators of diagenetic status in marine organic matter. Geochim Cosmochim Acta 61:5363–5369. doi: 10.1016/S0016-7037(97)00312-8. [DOI] [Google Scholar]
- 58.Dauwe B, Middelburg JJ. 2003. Amino acids and hexosamines as indicators of organic matter degradation state in North Sea sediments. Limnol Oceanogr 43:782–798. doi: 10.4319/lo.1998.43.5.0782. [DOI] [Google Scholar]
- 59.Orsi WD, Richards TA, Francis WR. 2018. Predicted microbial secretomes and their target substrates in marine sediment. Nat Microbiol 3:32–37. doi: 10.1038/s41564-017-0047-9. [DOI] [PubMed] [Google Scholar]
- 60.Baker BJ, Lazar CS, Teske AP, Dick GJ. 2015. Genomic resolution of linkages in carbon, nitrogen, and sulfur cycling among widespread estuary sediment bacteria. Microbiome 3:14. doi: 10.1186/s40168-015-0077-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Schmidt J. 2016. Microbial extracellular enzymes in marine sediments: methods development and potential activities in the Baltic Sea deep biosphere. M.Sc. thesis University of Tennessee, Knoxville, TN. [Google Scholar]
- 62.Lloyd KG, Schreiber L, Petersen DG, Kjeldsen KU, Lever MA, Steen AD, Stepanauskas R, Richter M, Kleindienst S, Lenk S, Schramm A, Jorgensen BB. 2013. Predominant archaea in marine sediments degrade detrital proteins. Nature 496:215–218. doi: 10.1038/nature12033. [DOI] [PubMed] [Google Scholar]
- 63.Marshall IPG, Starnawski P, Cupit C, Fernández Cáceres E, Ettema TJG, Schramm A, Kjeldsen KU. 2017. The novel bacterial phylum Calditrichaeota is diverse, widespread and abundant in marine sediments and has the capacity to degrade detrital proteins. Environ Microbiol Rep 9:397–403. doi: 10.1111/1758-2229.12544. [DOI] [PubMed] [Google Scholar]
- 64.Hedges JI, Keil RG, Benner R. 1997. What happens to terrestrial organic matter in the ocean? Org Geochem 27:195–212. doi: 10.1016/S0146-6380(97)00066-1. [DOI] [Google Scholar]
- 65.Seitz KW, Lazar CS, Hinrichs K-U, Teske AP, Baker BJ. 2015. Genomic reconstruction of a novel, deeply branched sediment archaeal phylum with pathways for acetogenesis and sulfur reduction. ISME J 10:1696. doi: 10.1038/ismej.2015.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Klippel B, Sahm K, Basner A, Wiebusch S, John P, Lorenz U, Peters A, Abe F, Takahashi K, Kaiser O, Goesmann A, Jaenicke S, Grote R, Horikoshi K, Antranikian G. 2014. Carbohydrate-active enzymes identified by metagenomic analysis of deep-sea sediment bacteria. Extremophiles 18:853–863. doi: 10.1007/s00792-014-0676-3. [DOI] [PubMed] [Google Scholar]
- 67.Thureborn P, Franzetti A, Lundin D, Sjöling S. 2016. Reconstructing ecosystem functions of the active microbial community of the Baltic Sea oxygen depleted sediments. PeerJ 4:e1593. doi: 10.7717/peerj.1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chung IK, Beardall J, Mehta S, Sahoo D, Stojkovic S. 2011. Using marine macroalgae for carbon sequestration: a critical appraisal. J Appl Phycol 23:877–886. doi: 10.1007/s10811-010-9604-9. [DOI] [Google Scholar]
- 69.Hoarau G, Coyer JA, Veldsink JH, Stam WT, Olsen JL. 2007. Glacial refugia and recolonization pathways in the brown seaweed Fucus serratus. Mol Ecol 16:3606–3616. doi: 10.1111/j.1365-294X.2007.03408.x. [DOI] [PubMed] [Google Scholar]
- 70.Ale MT, Mikkelsen JD, Meyer AS. 2011. Important determinants for fucoidan bioactivity: a critical review of structure-function relations and extraction methods for fucose-containing sulfated polysaccharides from brown seaweeds. Mar Drugs 9:2106–2130. doi: 10.3390/md9102106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Teplyuk AV, Samarov NI, Korzhenkov AA, Ulyanova MO, Goeva MA, Kublanov IV, Kanapatskii TA, Pimenov NV, Toshchakov SV. 2017. Analysis of chitinase diversity in the Baltic Sea bottom sediments. Microbiology 86:150–154. doi: 10.1134/S0026261717010131. [DOI] [PubMed] [Google Scholar]
- 72.Wu L, Kellogg L, Devol AH, Tiedje JM, Zhou J. 2008. Microarray-based characterization of microbial community functional structure and heterogeneity in marine sediments from the Gulf of Mexico. Appl Environ Microbiol 74:4516–4529. doi: 10.1128/AEM.02751-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Boyer JN. 1994. Aerobic and anaerobic degradation and mineralization of 14C-chitin by water column and sediment inocula of the York River estuary, Virginia. Appl Environ Microbiol 60:174–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Gooday GW. 1990. The ecology of chitin degradation, p 387–430. In Marshall KC.(ed), Advances in microbial ecology. Springer US, Boston, MA. [Google Scholar]
- 75.Glombitza C, Jaussi M, Røy H, Seidenkrantz M-S, Lomstein BA, Jørgensen BB. 2015. Formate, acetate, and propionate as substrates for sulfate reduction in sub-arctic sediments of Southwest Greenland. Front Microbiol 6:27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lamed R, Zeikus JG. 1980. Ethanol production by thermophilic bacteria: relationship between fermentation product yields of and catabolic enzyme activities in Clostridium thermocellum and Thermoanaerobium brockii. J Bacteriol 144:569–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lamed R, Zeikus JG. 1980. Glucose fermentation pathway of Thermoanaerobium brockii. J Bacteriol 141:1251–1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Leskovac V, Trivić S, Peričin D. 2002. The three zinc‐containing alcohol dehydrogenases from baker's yeast, Saccharomyces cerevisiae. FEMS Yeast Res 2:481–494. doi: 10.1016/S1567-1356(02)00157-5. [DOI] [PubMed] [Google Scholar]
- 79.Kirchman DL, Hanson TE, Cottrell MT, Hamdan LJ. 2014. Metagenomic analysis of organic matter degradation in methane-rich Arctic Ocean sediments. Limnol Oceanogr 59:548–559. doi: 10.4319/lo.2014.59.2.0548. [DOI] [Google Scholar]
- 80.Zhuang G-C, Lin Y-S, Elvert M, Heuer VB, Hinrichs K-U. 2014. Gas chromatographic analysis of methanol and ethanol in marine sediment porewaters: validation and implementation of three pretreatment techniques. Mar Chem 160:82–90. doi: 10.1016/j.marchem.2014.01.011. [DOI] [Google Scholar]
- 81.Graue J, Engelen B, Cypionka H. 2011. Degradation of cyanobacterial biomass in anoxic tidal-flat sediments: a microcosm study of metabolic processes and community changes. ISME J 6:660–669. doi: 10.1038/ismej.2011.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Friberg YB. 2015. The paleoceanography of Kattegat during the last deglaciation from benthic foraminiferal stable isotopes. Master thesis Lund University, Lund, Sweden. [Google Scholar]
- 83.Orsi WD, Coolen MJL, Wuchter C, He L, More KD, Irigoien X, Chust G, Johnson C, Hemingway JD, Lee M, Galy V, Giosan L. 2017. Climate oscillations reflected within the microbiome of Arabian Sea sediments. Sci Rep 7:6040. doi: 10.1038/s41598-017-05590-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Vuillemin A, Ariztegui D, Horn F, Kallmeyer J, Orsi WD, the PASADO Science Team. 2018. Microbial community composition along a 50 000-year lacustrine sediment sequence. FEMS Microbiol Ecol 94:fiy029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Inagaki F, Hinrichs KU, Kubo Y, Bowles MW, Heuer VB, Hong WL, Hoshino T, Ijiri A, Imachi H, Ito M, Kaneko M, Lever MA, Lin YS, Methé BA, Morita S, Morono Y, Tanikawa W, Bihan M, Bowden SA, Elvert M, Glombitza C, Gross D, Harrington GJ, Hori T, Li K, Limmer D, Liu CH, Murayama M, Ohkouchi N, Ono S, Park YS, Phillips SC, Prieto-Mollar X, Purkey M, Riedinger N, Sanada Y, Sauvage J, Snyder G, Susilawati R, Takano Y, Tasumi E, Terada T, Tomaru H, Trembath-Reichert E, Wang DT, Yamada Y. 2015. Exploring deep microbial life in coal-bearing sediment down to 2.5 km below the ocean floor. Science 349:420–424. doi: 10.1126/science.aaa6882. [DOI] [PubMed] [Google Scholar]
- 86.Seeberg-Elverfeldt J, Schlüter M, Feseker T, Kölling M. 2005. Rhizon sampling of porewaters near the sediment/water interface of aquatic systems. Limnol Oceanogr Methods 3:361–371. doi: 10.4319/lom.2005.3.361. [DOI] [Google Scholar]
- 87.Manheim FT. 1966. A hydraulic squeezer for obtaining interstitial water from consolidated and unconsolidated sediments. US Geological Survey, Reston, VA. [Google Scholar]
- 88.Røy H, Weber HS, Tarpgaard IH, Ferdelman TG, Jørgensen BB. 2014. Determination of dissimilatory sulfate reduction rates in marine sediment via radioactive 35S tracer. Limnol Oceanogr Methods 12:196–211. doi: 10.4319/lom.2014.12.196. [DOI] [Google Scholar]
- 89.Vineis JH, Ringus DL, Morrison HG, Delmont TO, Dalal S, Raffals LH, Antonopoulos DA, Rubin DT, Eren AM, Chang EB, Sogin ML. 2016. Patient-specific Bacteroides genome variants in pouchitis. mBio 7:e01713-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Tully BJ, Graham ED, Heidelberg JF. 2018. The reconstruction of 2,631 draft metagenome-assembled genomes from the global oceans. Sci Data 5:170203. doi: 10.1038/sdata.2017.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Müller O, Bang-Andreasen T, White RA III, Elberling B, Taş N, Kneafsey T, Jansson JK, Øvreas L. 2018. Disentangling the complexity of permafrost soil by using high resolution profiling of microbial community composition, key functions and respiration rates. Environ Microbiol 20:4328–4342. doi: 10.1111/1462-2920.14348. [DOI] [PubMed] [Google Scholar]
- 92.Singleton CM, McCalley CK, Woodcroft BJ, Boyd JA, Evans PN, Hodgkins SB, Chanton JP, Frolking S, Crill PM, Saleska SR, Rich VI, Tyson GW. 2018. Methanotrophy across a natural permafrost thaw environment. ISME J 12:2544–2558. doi: 10.1038/s41396-018-0065-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Petersen KR, Streett DA, Gerritsen AT, Hunter SS, Settles ML. 2015. Super deduper, fast PCR duplicate detection in fastq files . 6th ACM Conference on Bioinformatics, Computational Biology and Health Informatics, 9 to 12 September 2015, Atlanta, GA. [Google Scholar]
- 94.Li D, Liu C-M, Luo R, Sadakane K, Lam T-W. 2015. MEGAHIT: an ultra-fast single-node solution for large and complex metagenomics assembly via succinct de Bruijn graph. Bioinformatics 31:1674–1676. doi: 10.1093/bioinformatics/btv033. [DOI] [PubMed] [Google Scholar]
- 95.Li D, Luo R, Liu C-M, Leung C-M, Ting H-F, Sadakane K, Yamashita H, Lam T-W. 2016. MEGAHIT v1.0: a fast and scalable metagenome assembler driven by advanced methodologies and community practices. Methods 102:3–11. doi: 10.1016/j.ymeth.2016.02.020. [DOI] [PubMed] [Google Scholar]
- 96.Eren AM, Esen ÖC, Quince C, Vineis JH, Morrison HG, Sogin ML, Delmont TO. 2015. Anvi’o: an advanced analysis and visualization platform for ‘omics data. PeerJ 3:e1319. doi: 10.7717/peerj.1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods 9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Hyatt D, Chen G-L, LoCascio PF, Land ML, Larimer FW, Hauser LJ. 2010. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics 11:119. doi: 10.1186/1471-2105-11-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Jones P, Binns D, Chang HY, Fraser M, Li W, McAnulla C, McWilliam H, Maslen J, Mitchell A, Nuka G, Pesseat S, Quinn AF, Sangrador-Vegas A, Scheremetjew M, Yong SY, Lopez R, Hunter S. 2014. InterProScan 5: genome-scale protein function classification. Bioinformatics 30:1236–1240. doi: 10.1093/bioinformatics/btu031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Finn RD, Bateman A, Clements J, Coggill P, Eberhardt RY, Eddy SR, Heger A, Hetherington K, Holm L, Mistry J, Sonnhammer ELL, Tate J, Punta M. 2013. Pfam: the protein families database. Nucleic Acids Res 42:D222–D230. doi: 10.1093/nar/gkt1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Haft DH, Selengut JD, Richter RA, Harkins D, Basu MK, Beck E. 2012. TIGRFAMs and genome properties in 2013. Nucleic Acids Res 41:D387–D395. doi: 10.1093/nar/gks1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Petersen TN, Brunak S, Heijne von G, Nielsen H. 2011. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods 8:785–786. doi: 10.1038/nmeth.1701. [DOI] [PubMed] [Google Scholar]
- 103.Yu NY, Wagner JR, Laird MR, Melli G, Rey S, Lo R, Dao P, Sahinalp SC, Ester M, Foster LJ, Brinkman FSL. 2010. PSORTb 3.0: improved protein subcellular localization prediction with refined localization subcategories and predictive capabilities for all prokaryotes. Bioinformatics 26:1608–1615. doi: 10.1093/bioinformatics/btq249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Kanehisa M, Sato Y, Kawashima M, Furumichi M, Tanabe M. 2016. KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res 44:D457–D462. doi: 10.1093/nar/gkv1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Caspi R, Altman T, Billington R, Dreher K, Foerster H, Fulcher CA, Holland TA, Keseler IM, Kothari A, Kubo A, Krummenacker M, Latendresse M, Mueller LA, Ong Q, Paley S, Subhraveti P, Weaver DS, Weerasinghe D, Zhang P, Karp PD. 2013. The MetaCyc database of metabolic pathways and enzymes and the BioCyc collection of Pathway/Genome Databases. Nucleic Acids Res 42:D459–D471. doi: 10.1093/nar/gkt1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Rawlings ND, Barrett AJ. 1993. Evolutionary families of peptidases. Biochem J 290:205–218. doi: 10.1042/bj2900205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Buchfink B, Xie C, Huson DH. 2014. Fast and sensitive protein alignment using DIAMOND. Nat Methods 12:59–60. doi: 10.1038/nmeth.3176. [DOI] [PubMed] [Google Scholar]
- 108.Huson DH, Mitra S, Ruscheweyh H-J, Weber N, Schuster SC. 2011. Integrative analysis of environmental sequences using MEGAN4. Genome Res 21:1552–1560. doi: 10.1101/gr.120618.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Huson DH, Beier S, Flade I, Górska A, El-Hadidi M, Mitra S, Ruscheweyh H-J, Tappu R. 2016. MEGAN community edition–interactive exploration and analysis of large-scale microbiome sequencing data. PLoS Comput Biol 12:e1004957. doi: 10.1371/journal.pcbi.1004957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Dixon P. 2003. VEGAN, a package of R functions for community ecology. J Veg Sci 14:927–930. doi: 10.1111/j.1654-1103.2003.tb02228.x. [DOI] [Google Scholar]
- 111.Papadomanolaki NM, Dijkstra N, van Helmond NAGM, Hagens M, Bauersachs T, Kotthoff U, Sangiorgi F, Slomp CP. 2018. Controls on the onset and termination of past hypoxia in the Baltic Sea. Palaeogeogr Palaeoclimatol Palaeoecol 490:347–354. doi: 10.1016/j.palaeo.2017.11.012. [DOI] [Google Scholar]
- 112.Hardisty DS, Riedinger N, Planavsky NJ, Asael D, Andrén T, Jørgensen BB, Lyons TW. 2016. A Holocene history of dynamic water column redox conditions in the Landsort Deep, Baltic Sea. Am J Sci 316:713–745. doi: 10.2475/08.2016.01. [DOI] [Google Scholar]
- 113.Wickham H. 2011. ggplot2: elegant graphics for data analysis. Biometrics 67:678–679. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Metagenomic data for M59E-0.25m, M59E-15m, M65C-0.25m, M65C-3mbsf, and M65C-10mbsf can be found in the National Center for Biotechnology Information (NCBI) under BioProject number PRJNA433242. Metagenomes from all other samples here were sequenced as described by Marshall et al. (36) and were retrieved from the NCBI SRA project SRP068645. Metatranscriptomes were sequenced as described by Zinke et al. (37) and archived under the NCBI SRA project SRP108285.