Abstract
It is clear that alcohol consumption is a major risk factor in the pathogenesis of head and neck squamous cell carcinoma (HNSCC); however, the molecular mechanism underlying the pathogenesis of alcohol-associated HNSCC remains poorly understood. The aim of the present study was to identify and characterize P-element-induced wimpy testis (PIWI)-interacting RNAs (piRNAs) and PIWI proteins dysregulated in alcohol-associated HNSCC to elucidate their function in the development of this cancer. Using next generation RNA-sequencing (RNA-seq) data obtained from 40 HNSCC patients, the piRNA and PIWI protein expression of HNSCC samples was compared between alcohol drinkers and non-drinkers. A separate piRNA expression RNA-seq analysis of 18 non-smoker HNSCC patients was also conducted. To verify piRNA expression, reverse transcription-quantitative polymerase chain reaction (RT-qPCR) was performed on the most differentially expressed alcohol-associated piRNAs in ethanol and acetaldehyde-treated normal oral keratinocytes. The correlation between piRNA expression and patient survival was analyzed using Kaplan-Meier estimators and multivariate Cox proportional hazard models. A comparison between alcohol drinking and non-drinking HNSCC patients demonstrated that a panel of 3,223 piRNA transcripts were consistently detected and differentially expressed. RNA-seq analysis and in vitro RT-qPCR verification revealed that 4 of these piRNAs, piR-35373, piR-266308, piR-58510 and piR-38034, were significantly dysregulated between drinking and non-drinking cohorts. Of these four piRNAs, low expression of piR-58510 and piR-35373 significantly correlated with improved patient survival. Furthermore, human PIWI-like protein 4 was consistently upregulated in ethanol and acetaldehyde-treated normal oral keratinocytes. These results demonstrate that alcohol consumption may cause dysregulation of piRNA expression in HNSCC and in vitro verifications identified 4 piRNAs that may be involved in the pathogenesis of alcohol-associated HNSCC.
Keywords: head and neck squamous cell carcinoma, piwi-interacting RNA, alcohol, epigenetics
Introduction
Head and neck squamous cell carcinoma (HNSCC) is the sixth most common type of cancer worldwide and despite overall advances in cancer treatment, mortality rates of HNSCC have not improved significantly in the past three decades (1,2). Early diagnosis of HNSCC results in a 5-year survival rate of 75%; however, the majority of HNSCC patients present with metastatic disease at the time of diagnosis and thus exhibit a 5-year survival rate of 35% (3). Therefore, it is critical to understand the molecular basis of HNSCC pathogenesis and identify the early genetic and epigenetic changes to enable early detection and diagnosis of HNSCC.
Alcohol consumption, in addition to other factors, including tobacco use and human papilloma virus infection, is a well-established major risk factor for HNSCC (4). Over 75% of HNSCC cases are caused by alcohol consumption and/or tobacco use (5). Although both habits are commonly linked, alcohol consumption has also been identified as an independent risk factor for HNSCC (6,7). However, the mechanism by which alcohol leads to cellular transformation and the molecular basis of this risk remains unclear.
Recently, the field of epigenetic research has gained increasing attention as it has been revealed that alterations in the epigenetic landscape play a role in HNSCC pathogenesis (8,9). More specifically, non-coding RNAs (ncRNAs) are considered critical in the development of many diseases, including cancer (10). In 2006, a novel group of ncRNAs, termed P-element-induced wimpy testis (PIWI)-interacting RNAs (piRNAs), was identified in mammalian germ line cells (11). piRNAs are 24–30 nucleotides in length and associate with PIWI-class Argonaute proteins (12,13). Their main biological functions include PIWI-dependent transposon silencing, heterochromatin modification, and germ cell maintenance (14). As cancer cells and germ cells share a number of biological features, such as rapid proliferation and an undifferentiated state, piRNAs are currently being investigated in the context of cancer. Several studies have demonstrated that piRNAs exhibit both oncogenic and tumor suppressive functions in a variety of cancer types, including gastric, breast, and colon cancer (14–17).
The newly identified piRNAs are involved in a myriad of gene regulation functions and therefore present a promising field of study. Currently, their precise function in carcinogenesis is unclear; however, this novel class of ncRNAs may present potential therapeutic targets or prognostic biomarker for early detection. To increase current knowledge of piRNAs, the present study aimed to determine the effects of alcohol consumption on piRNAs expression in HNSCC. Using RNA-sequencing (RNA-seq) analysis of HNSCC tissue samples obtained from alcohol drinkers and non-drinkers, we aimed to identify a panel of differentially expressed piRNAs between these two cohorts. Verification of these alcohol-associated piRNAs in ethanol treated normal oral keratinocyte cells allows us to determine the piRNAs potentially involved in the early events of HNSCC pathogenesis.
Materials and methods
piRNA expression analysis
External RNA-seq datasets and corresponding clinical information of 40 HNSCC patients were obtained from The Cancer Genome Atlas (TCGA) (https://tcga-data.nci.nih.gov), which contained RNASeq or RNASeqv2 HNSCC data. Each of the datasets obtained was aligned to a human reference genome, (hg19) using Tophat (https://ccb.jhu.edu/software/tophat/index.shtml) (18) and piRNA transcript assembly was performed using Cufflinks (http://cole-trapnell-lab.github.io/cufflinks/cuffdiff/) (18). Differential expression analysis was performed using Cuffdiff, the Cufflinks transcript quantification engine (18). piRNAs with extremely low expression [Fragments Per Kilobase of transcript per Million mapped reads (FPKM) of <1 in >50% of the samples] were excluded from the analysis. piRNAs with expression that differed by a fold change >1 in magnitude (P<0.05) between drinkers and non-drinkers were identified by Cuffdiff to be differentially expressed. To generate a more selective panel of piRNAs for further analysis, more stringent filtering of the piRNAs was performed based on differential expression levels that were >two-fold (P<0.05). TCGA obtained written informed consent from all patients prior to publishing datasets.
piRNA survival analysis
All clinical data used in the analysis was obtained from TCGA. For survival analysis, piRNA expression was modeled as a binary variable, with the ‘low expression’ group corresponding to half of HNSCC patients with expression below the median, and ‘high expression’ corresponding to the remaining half of patients with expression above the median. For multivariate Cox analysis, covariates included patient age, clinical stage, pathologic stage, sex and tumor grade. The statsoft STATISTICA software version 12 was used for all survival analyses.
Cell lines and cell culture
Two normal oral keratinocyte cell lines, OKF4 and OKF6, were provided by the Rheinwald Lab at Harvard Medical School (Boston, MA, USA). The oral keratinocytes were cultured in keratinocyte serum-free medium (SFM; 1X) with L-glutamine, supplemented with 0.2 ng/ml human recombinant epidermal growth factor (EGF) 1–53 (recombinant 1–53 of EGF), 25 µg/ml bovine pituitary extract (BPE), 0.3 mM CaCl2 and penicillin streptomycin (Thermo Fisher Scientific, Inc., Waltham, MA, USA). At 30% confluence, the cells were cultured at 37°C and at 5% CO2 for 3 weeks with equal parts keratinocyte-SFM medium and DFK. DFK medium was made with equal parts Dulbecco's modified Eagle's medium and F-12 (Corning Inc., Corning, New York, NY, USA), and supplemented with 0.2 ng/ml EGF 1–53, 25 µg/ml BPE, 2 mM L-glutamine, and 10 ml penicillin streptomycin (10,000 units/ml penicillin, 10,000 ug/ml streptomycin).
Ethanol and acetaldehyde treatment
Oral keratinocyte cell lines OKF4 and OKF6 were plated on 60×15 mm non-pyrogenic, non-cytotoxic plates (SARSTEDT, North Rhine-Westphalia, Nümbrecht, Germany) at 10% confluence. After reaching >30% confluence, they were then treated with increasing doses of ethanol and acetaldehyde to simulate the effects of alcohol consumption on human oral epithelial cells prior to carcinogenesis. Clinically relevant ethanol concentrations of 0.1, 0.3, and 1% (v/v) were selected to replicate the alcohol levels of social, moderate and heavy drinkers, respectively. Long term alcohol consumption was simulated via daily treatment with ethanol diluted in oral keratinocyte-SFM (Thermo Fisher Scientific, Inc.) for 28 days. Ethanol and growth medium were replaced daily and plates were sealed with paraffin film to prevent ethanol evaporation.
Oral keratinocyte cell lines OKF4 an OKF6 were also treated with acetaldehyde (Sigma-Aldrich; Merck KGaA, Darmstadt, Germany), the first metabolite of ethanol and a potential carcinogen. Physiologically relevant doses of acetaldehyde were used, as determined by saliva concentrations in alcohol consumers (19). Therefore, cells were treated with 75, 150 and 300 µm acetaldehyde diluted in growth medium for 48 h. Acetaldehyde and growth media were replaced every 4 h. Due to the volatility and short half-life of acetaldehyde, long-term treatment was not possible and thus had to be administered every 4 h. Cell lines were passaged to 30–80% confluence prior to harvesting RNA for subsequent experiments.
Verification of differential expression by reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
Approximately 45 µl of total RNA was extracted from cultured cells using the RNeasy Mini kit (Qiagen, Hilden, Germany). cDNA was then synthesized using reverse transcriptase (Invitrogen; Thermo Fisher Scientific, Inc.), according to the manufacturer's instructions. Quantitative PCR was performed using SYBR-Green Reagent (Roche Diagnostics, Basel, Switzerland), Ultrapure Water (RX Biosciences; Gaithersburg, MD, USA), as well as the necessary cDNA and primers. The reaction was run using the Applied Biosystems 7300 Real-Time PCR System (Applied Biosystems; Thermo Fisher Scientific, Inc.) which ran a 2 h PCR consisting of a 20 sec holding stage where the reaction is held at 95°C, followed by a cycling stage consisting of 3 sec at 95°C, then 30 sec at 60°C. This cycling stage was repeated 40 times before concluding with a melt curve stage, where the reaction is held at 95°C for 15 sec, 60°C for 1 min, then 95°C again for another 15 sec. Experiments were performed in triplicate and relative expression of piRNAs was calculated and normalized using the 2−ΔΔCt method (20) relative to U6 control. Primers were designed and synthesized by Eurofins Genomics (Huntsville, AL, USA). The primer sequences were as follows:
piR-258904, 5′-TGAGAACATTGGATTTTTCA-3′; piR-35373, 5′-TCCACACACCAAGCCTTGGA-3′; piR-266308, 5′-ACGGCCCTGGCGGAGCGCAA-3′; piR-34946, 5′-TCATGAGGTCAGGAGTTCAA-3′; piR-58510, 5′-GCCTGGCCAACATGGCAAAA-3′; piR-43219, 5′-CACCTTGGGAGGCCAA-3′; piR-38034, 5′-TCGCTTGAACCTGGAA-3′ and piR-70732, 5′-TGATGATGGTGGTGGTGGTGTCGGTGA-3′U6 Human forward control primer, 5-CGCAAGGATGACACGCAAATTC-3′.
The forward and universal reverse primer were both provided in the QuantiMir RT kit Small RNA Quantitation System (System Biosciences, Palo Alto, CA, USA) and were used as per manufacturer's instruction, the sequence to the reverse primer is commercialized and thus not available.
Statistical analysis
The two-tailed student's t test was used to test for statistical significance between the two cohorts in every in vitro analysis as well as survival tests. The significance of the tests in the differential and survival analyses was automatically calculated by the software provided, which were Statsoft by STATISTICA and Cuffdiff maintained by the Trapnell Lab. P<0.05 was considered to indicate a statistically significant difference.
Results
Identification of differentially expressed piRNAs associated with alcohol consumption
RNA-seq data was analyzed to determine the difference in piRNA expression between alcohol drinker and non-drinker HNSCC patients using data obtained from TCGA. The patient cohort included 20 alcohol drinkers and 20 non-drinkers with equal numbers of smoker and non-smokers in each group (Table I). The primary sequence data files of each patient in the cohort were downloaded from the controlled access data tier of the Cancer Genomics Hub (https://cghub.ucsc.edu/). The data files were then mapped to a piRNA reference using Tophat and transcripts were assembled using Cufflinks. Based on the normalized FPKM output, the differential expression of 3,223 piRNA transcripts was compared between the alcohol drinkers and non-drinkers (Fig. 1). From the expressed transcripts, piRNAs that exhibited a fold change >2 (P<0.05) between alcohol drinkers and non-drinkers were considered to be differentially expressed. Using these parameters, upregulation of piR-58510, −43219, −38034, −34946 and downregulation of piR-70732 was identified in alcohol drinkers (Table II). In order to determine the piRNAs independently affected by alcohol consumption alone, the same analysis was performed in the non-smoking patient cohort. The results revealed an upregulation of piR-258904, −35373, −266308 and −34946 in the alcohol drinking, non-smoking cohort.
Table I.
Demographics and clinicopathological characteristics of 40 head and neck squamous cell carcinoma patients.
| Non-drinkers (n=20 | Drinkers (n=20) | ||||
|---|---|---|---|---|---|
| Parameter | Total patients n (%) | Smokers n (%) | Non-smokers n (%) | Smokers n (%) | Non-smokers n (%) |
| Sex | |||||
| Male | 28 (70) | 6 (55) | 5 (56) | 10 (91) | 7 (78) |
| Female | 12 (30) | 5 (45) | 4 (44) | 1 (9) | 2 (22) |
| Alcoholic drinks per day, n=0 | |||||
| 20 (50) | N/A | N/A | 0 (0) | 0 (0) | |
| 1–2 | 5 (13) | N/A | N/A | 0 (0) | 5 (56) |
| >2 | 15 (37) | N/A | N/A | 11 (100) | 4 (44) |
| Vital status | |||||
| Deceased | 32 (80) | 7 (64) | 8 (89) | 9 (82) | 8 (89) |
| Alive | 8 (20) | 4 (36) | 1 (11) | 2 (18) | 1 (11) |
| Tumor site | |||||
| Oral | 24 (60) | 8 (73) | 6 (67) | 6 (55) | 4 (44) |
| Pharyneal | 10 (25) | 1 (9) | 3 (33) | 1 (9) | 5 (56) |
| Laryngeal | 6 (15) | 2 (18) | 0 (0) | 4 (36) | 0 (0) |
| Tumor stagea | |||||
| I/II | 9 (23) | 2 (20) | 1 (11) | 3 (27) | 3 (33) |
| III/IV | 30 (77) | 8 (80) | 8 (89) | 8 (73) | 6 (67) |
| Tumor grade | |||||
| GX | 1 (2) | 0 (0) | 0 (0) | 0 (0) | 1 (11) |
| G1-G2 | 30 (75) | 10 (91) | 7 (78) | 8 (73) | 5 (56) |
| G3-G4 | 9 (23) | 1 (9) | 2 (22) | 3 (27) | 3 (33) |
Data for 1 patient of the non-drinker/non-smoker group was not available. All patient data was obtained from The Cancer Genome Atlas. N/A, not applicable.
Figure 1.
Analysis of RNA-sequencing data of HNSCC samples. (A) Heatmap representing the piRNA FPKMs of the entire patient cohort (n=40). Patients were divided into drinking and non-drinking cohorts and further subdivided by tobacco usage. Only piRNAs that were expressed in >20 patients are shown. (B) Kaplan Meier curves indicate that low expression of piR-58510 and piR-35373 is associated with longer survival times. Log rank tests were performed to determine the P-values shown. (C) Cox proportional hazard ratios further demonstrate the correlation between piRNA-58510 and piR-35373 expression and overall prognosis. The hazard ratios presented for both univariate and multivariate tests are based on the low expression of each piRNA. Vertical lines represent censored patients. HNSCC, head and neck squamous cell carcinoma; PIWI, P-element-induced wimpy testis; piRNA, PIWI-interacting RNA; FPKM, Fragments per Kilobase of transcript per Million mapped reads; HR, hazard ratio; CI, confidence interval.
Table II.
Differentially expressed piRNAs and their corresponding fold changes in expression.
| piRNA | Fold change | P-value | Sequence (5′-3′) |
|---|---|---|---|
| Non-smokersa | |||
| piR-258904 | 3.26275255 | 0.031 | TGAGAACATTGGATTTTTCACCCTAG |
| piR-35373 | 2.76213293 | 0.031 | TCCACACACCAAGCCTTGGATTGGTCTGG |
| piR-266308 | 2.57372618 | 0.019 | ACGGCCCTGGCGGAGCGC |
| piR-34946 | 2.29238305 | 0.027 | TCATGAGGTCAGGAGTTCAAGACCAGCC |
| All patientsb | |||
| piR-58510 | 5.242446202 | 0.0048 | GCCTGGCCAACATGGCAAAACGTCGTGTCTA |
| piR-34946 | 3.446885753 | 0.0014 | TCATGAGGTCAGGAGTTCAAGACCAGCC |
| piR-43219 | 3.389853452 | 0.0032 | CACCTTGGGAGGCCAAGGCAGGCAGATCAT |
| piR-38034 | 3.316586523 | 0.0068 | TCGCTTGAACCTGGAAGGCGGAGGTTGC |
| piR-70732 | 0.359078784 | 0.057 | TGATGATGGTGGTGGTGGTGTCGGTGA |
piRNA expression was compared between alcohol drinkers and non-drinkers in the non-smoking cohort (n=18).
piRNA expression was compared between alcohol drinkers and non-drinkers in all patients (n=40). piRNA, PIWI-interacting RNA; PIWI, P-element-induced wimpy testis.
Expression of piR-58510 and piR-35373 correlates with better patient survival
Using the available clinical data for the entire patient cohort, the association between each differentially expressed piRNA and patient survival time was analyzed. The patient cohort was divided into high and low piRNA expression groups for univariate and multivariate Cox proportional hazard test analysis (adjusted for age and sex). Testing the 8 piRNAs found to be dysregulated in the RNA-seq analysis (−58510, −43219, −38034, −34946, −258904, −35373, −266308), revealed that only piR-58510 and piR-35373 exhibited a significant correlation to survival (P<0.05) (Fig. 1B and C). These results indicated that low expression of piR-58510 and piR-35373 correlates with longer patient survival times.
In vitro ethanol exposure leads to dysregulation of identified piRNAs in normal oral keratinocytes
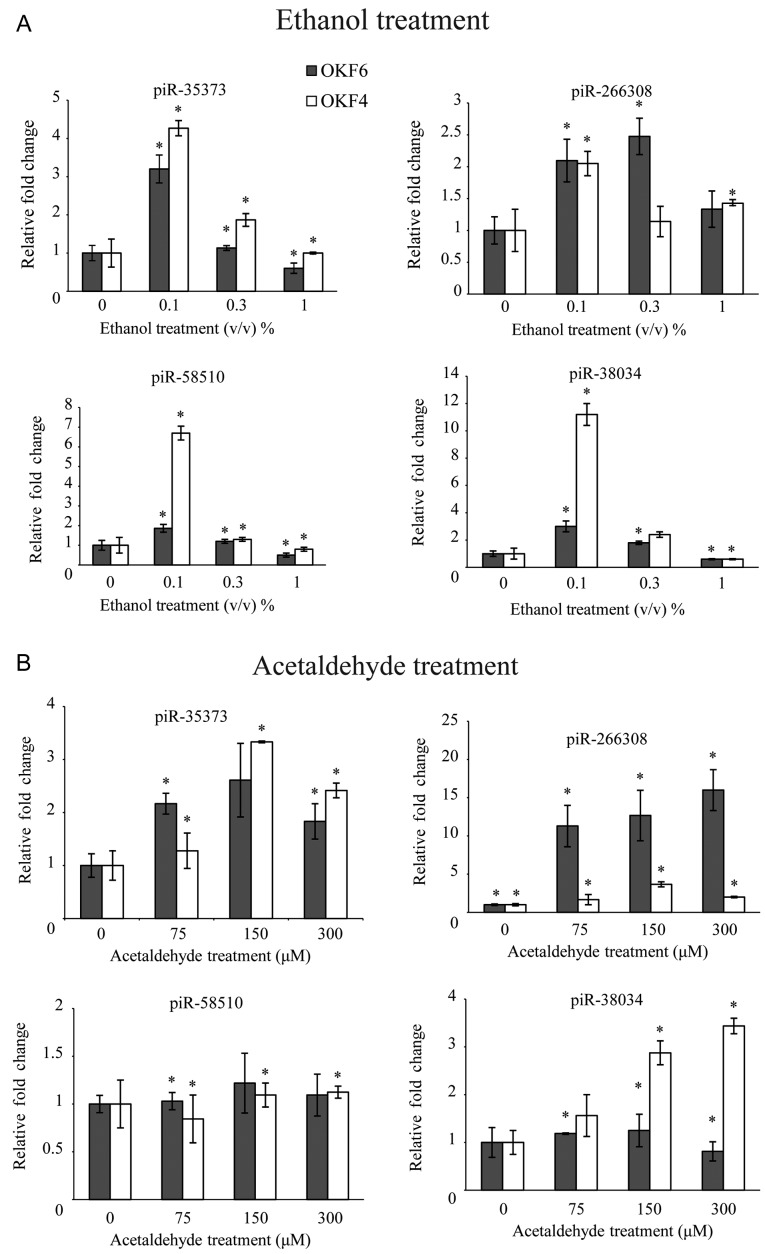
To validate that the clinically relevant piRNAs identified were associated with alcohol consumption, in vitro ethanol treatment using early passage oral epithelial culture cells OKF4 and OKF6 was performed. Normal oral epithelial cells were selected in order to investigate whether alcohol causes malignant transformation via dysregulation of the identified piRNAs. To mimic alcohol consumption, cell cultures were exposed to 0.1, 0.3, 1% ethanol daily for 4 weeks to simulate the alcohol levels of social, moderate and heavy drinkers, respectively. A preliminary MTS assay was performed to demonstrate that the selected doses did not cause significant cell toxicity (data not shown). Following treatment, RT-qPCR assays were performed to analyze the expression of each of the piRNAs identified from analysis of the clinical data. The data demonstrated that ethanol caused an upregulation of piR-35373, −266308, −58510, and −38034, which was consistent with the results obtained in the analysis of the RNA-seq data (Fig. 2A). Notably, the highest upregulation of these piRNAs occurred following treatment with 0.1% ethanol.
Figure 2.
Long term ethanol exposure and acetaldehyde treatment leads to dysregulation of identified piRNAs in vitro. Quantitative reverse-transcription polymerase chain reaction demonstrated that demonstrate that (A) ethanol and (B) acetaldehyde treatment of normal OKF4 and OKF6 oral epithelial cell cultures increases piRNA 35373, −266308, −58510 and −38034 in both cell lines. Data are presented as the mean ± standard deviation. PIWI, P-element-induced wimpy testis; piRNA, PIWI-interacting RNA. *P<0.05 vs. control.
In vitro acetaldehyde exposure leads to dysregulation of identified piRNAs in normal oral keratinocytes
Although alcohol consumption is a major risk factor for HNSCC, ethanol alone is not considered a carcinogen. However, acetaldehyde, the first metabolite of ethanol is a known carcinogen. To further validate that the candidate piRNAs (piR-35373, −266308, −58510 and −38034) are involved in alcohol-associated HNSCC, cells were treated with physiologically relevant doses of acetaldehyde for 48 h. A significant upregulation in piR-35373, piR-266308 and piR-38034 was identified in both OKF4 and OKF6 cell lines (P<0.05), with dose dependence stopping the effects at 300 mM, where high cell death is suspected to have affected expression of the piRNAs. The upregulation of those three pi-RNAs was similar to that observed with in vitro ethanol treatment. However, no significant differences in the expression levels of piR-58510 were identified in OKF4 or OKF6 cells following treatment with acetaldehyde (Fig. 2B).
Ethanol and acetaldehyde exposure leads to dysregulation of PIWI-like protein 4 (PIWIL4)
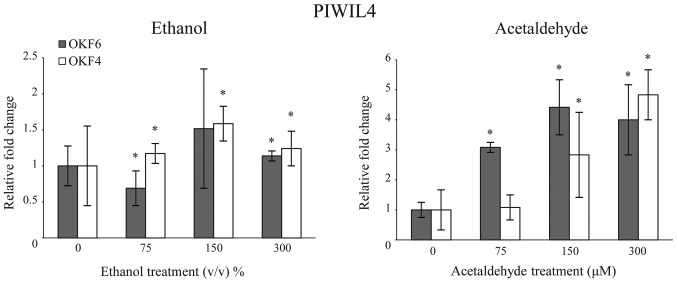
Although the role of piRNAs in cancer development remains unclear, their binding proteins are well-characterized and the 4 PIWI proteins expressed in humans have been demonstrated to exhibit functional involvement in a variety of cancer types (14). Of the 4 PIWI proteins identified (PIWIL1, PIWIL2, PIWIL3, PIWIL4), only the relative expression level of PIWIL4 was dysregulated in cells exposed to both ethanol and acetaldehyde treatments (Fig. 3).
Figure 3.
Long term ethanol exposure and acetaldehyde treatment leads to dysregulation of PIWIL4 in vitro. Quantitative reverse transcription-polymerase chain reaction demonstrated that ethanol and acetaldehyde treatment of normal OKF4 and OKF6 oral epithelial cell cultures increases the expression of PIWIL4 in both cell lines. Error bars represent standard deviation. Data are presented as the mean ± standard deviation. *P<0.05 vs. control. PIWI, P-element-induced wimpy testis; PIWIL4, PIWI-like protein 4.
Discussion
Alcohol consumption has been identified as a risk factor for the pathogenesis of HNSCC. Despite recent advances in HNSCC treatment, few studies has been published regarding the epigenetic biomarkers of alcohol-associated HNSCC (8), and survival rates for the disease have exhibited little improvement in the past three decades (21). The importance of epigenetics in alcohol-associated HNSCC remains unclear and elucidating its function may lead to the development of novel diagnostic and therapeutic targeting strategies (22).
To the best of our knowledge, the present study is the first to investigate the expression of piRNAs in HNSCCs. In addition, to date no studies have analyzed the effects of environmental factors or substance use on the expression of piRNAs and the present study is the first to demonstrate a dysregulation of piRNA expression due to alcohol consumption. In this study, the expression of 23,439 piRNA transcripts in alcohol drinkers and non-drinkers were compared. Among the 3,223 piRNA transcripts consistently detected among the samples, piR-58510, −34946, −43219, −38034 and −70732 (Table II) were significantly differentially expressed in alcohol-drinking HNSCC patients (fold change of ≥2 in all patients), while piR-258904, −35373, −266308 and −34946 (Table II) were significantly differentially expressed in the non-smoker cohort. Notably, only piR-34946 was identified in the analysis of both cohorts, which suggests that the synergistic effect between alcohol and tobacco usage may affect piRNA expression, and that this piRNA may be involved in the effects of both major risk factors. Previous studies have revealed an association between alcohol consumption and tobacco exposure an epigenetic dysregulation of ncRNAs (23,24). Furthermore, ncRNA dysregulation has been implicated in a number of cancer types (12). To date, none of the piRNAs identified in the present study have been functionally characterized and thus, their involvement in cancer progression remains unclear. The 3,223 piRNA transcripts consistently detected among the clinical patient samples was significantly less than the increasing number of piRNAs that have already been identified. This suggests that cancer cells, or specifically HNSCC cells, may not express the same number of piRNAs as germ line cells (25,26).
In the present study, RNA-seq data of HNSCC clinical samples was used to determine the differential expression of all expressed piRNA transcripts and to identify a panel of significantly dysregulated piRNAs in the alcohol-drinkers when compared with non-drinkers. To identify piRNAs with the greatest potential functional significance, the correlation between the expression levels of candidate transcripts in HNSCC patients and overall survival was analyzed using univariate and multivariate Cox regression analyses. The results identified low expression of 2 piRNAs, piR-58510 and piR-35373, was significantly associated with improved survival. Low expression of piR-58510 and piR-35373 correlated with longer survival times indicating that these piRNAs may act as oncogenes and thus, may present potential prognostic markers. This is the first study to reveal a correlation between piRNA expression and improved patient prognosis, which highlights the clinical relevance of piRNAs. Furthermore, 4 previously uncharacterized piRNAs, piR-35373, piR-266308, piR-58510 and piR-38034, and a PIWI protein that are hypothesized to be involved in the early events of alcohol-associated HNSCC pathogenesis were identified in non-cancerous oral keratinocytes. Using blat, a BLAST-like alignment tool (http://genome.ucsc.edu), piR-35373 revealed a sense overlap with the mRNA transcript of 3-phosphoglycerate dehydrogenase (PHGDH), a serine metabolism-associated protein that is highly expressed in several breast cancer subtypes and invasive lobular carcinoma (27,28). piR-35373 may act as a transcriptional regulator of PHGDH expression; however, the underlying molecular mechanism remains to be elucidated. PIWIL4 is upregulated in human cervical cancer tissues compared with adjacent normal tissues and thus acts as an oncogene by promoting cell growth and invasion and by downregulating p14ARF and p53 (16). The in vitro data obtained in the present study indicates that the alcohol-associated pathogenesis of HNSCC may be initiated by the dysregulation of PIWIL4 and its associated piRNAs.
Although the exact molecular function of the piRNAs identified remains unclear, we hypothesize that these piRNAs contribute to carcinogenesis by regulating gene expression of certain oncogenes or tumor suppressors, as demonstrated in previous piRNA studies (14,16). We acknowledge that the relatively limited number of patient datasets available in the database and the fact that only in-vitro validation was performed provide possible limitations to this study. Despite these limitations, we strongly endorse that these results demonstrate that alcohol consumption causes dysregulation of piRNA expression in HNSCC samples and in vitro data identified 4 piRNAs that may potentially serve as therapeutic targets or biomarkers for early detection of HNSCC.
Acknowledgements
Not applicable.
Funding
The present study was supported by funding from the National Institutes of Health (grant no. DE023242 to WMO.) and the University of California Academic Senate grant to WMO.
Availability of data and materials
The datasets generated and/or analyzed during the current study are available in The Cancer Genome Atlas (https://portal.gdc.cancer.gov/projects/TCGA-HNSC).
Authors' contributions
WMO, MAS and JWR were involved in conceptualization of the study. WMO was responsible for funding acquisition. MAS, JK, SZK, PXL, HZ and MAY performed the experiments. WMO and JWR supervised the project. MAS, JK and PL collaborated in the analysis of the data and constructed the figures. MAS and WMO wrote the manuscript. MAS, WMO and PXL reviewed and edited the manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Parkin DM, Bray F, Ferlay J, Pisani P. Global cancer statistics, 2002. CA Cancer J Clin. 2005;55:74–108. doi: 10.3322/canjclin.55.2.74. [DOI] [PubMed] [Google Scholar]
- 2.Nemunaitis J, O'Brien J. Head and neck cancer: Gene therapy approaches. Part 1: Adenoviral vectors. Expert Opin Biol Ther. 2002;2:177–185. doi: 10.1517/14712598.2.2.177. [DOI] [PubMed] [Google Scholar]
- 3.Silveira NJ, Varuzza L, Machado-Lima A, Lauretto MS, Pinheiro DG, Rodrigues RV, Severino P, Nobrega FG, Head and Neck Genome Project GENCAPO. Silva WA, Jr, et al. Searching for molecular markers in head and neck squamous cell carcinomas (HNSCC) by statistical and bioinformatic analysis of larynx-derived SAGE libraries. BMC Med Genomics. 2008;1:56. doi: 10.1186/1755-8794-1-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Argiris A, Karamouzis MV, Raben D, Ferris RL. Head and neck cancer. Lancet. 2008;371:1695–1709. doi: 10.1016/S0140-6736(08)60728-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hashibe M, Brennan P, Benhamou S, Castellsague X, Chen C, Curado MP, Dal Maso L, Daudt AW, Fabianova E, Fernandez L, et al. Alcohol drinking in never users of tobacco, cigarette smoking in never drinkers, and the risk of head and neck cancer: Pooled analysis in the international head and neck cancer epidemiology consortium. J Natl Cancer Inst. 2007;99:777–789. doi: 10.1093/jnci/djk179. [DOI] [PubMed] [Google Scholar]
- 6.Bobo JK, Husten C. Sociocultural influences on smoking and drinking. Alcohol Res Health. 2000;24:225–232. [PMC free article] [PubMed] [Google Scholar]
- 7.Sturgis EM, Wei Q, Spitz MR. Descriptive epidemiology and risk factors for head and neck cancer. Semin Oncol. 2004;31:726–733. doi: 10.1053/j.seminoncol.2004.09.013. [DOI] [PubMed] [Google Scholar]
- 8.Jithesh PV, Risk JM, Schache AG, Dhanda J, Lane B, Liloglou T, Shaw RJ. The epigenetic landscape of oral squamous cell carcinoma. Br J Cancer. 2013;108:370–379. doi: 10.1038/bjc.2012.568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Worsham MJ, Chen KM, Ghanem T, Stephen JK, Divine G. Epigenetic modulation of signal transduction pathways in HPV-associated HNSCC. Otolaryngol Head Neck Surg. 2013;149:409–416. doi: 10.1177/0194599813490895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huang T, Alvarez A, Hu B, Cheng SY. Noncoding RNAs in cancer and cancer stem cells. Chin J Cancer. 2013;32:582–593. doi: 10.5732/cjc.013.10170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Girard A, Sachidanandam R, Hannon GJ, Carmell MA. A germline-specific class of small RNAs binds mammalian Piwi proteins. Nature. 2006;442:199–202. doi: 10.1038/nature04917. [DOI] [PubMed] [Google Scholar]
- 12.Esteller M. Non-coding RNAs in human disease. Nat Rev Genet. 2011;12:861–874. doi: 10.1038/nrg3074. [DOI] [PubMed] [Google Scholar]
- 13.Galasso M, Sana ME, Volinia S. Non-coding RNAs: A key to future personalized molecular therapy? Genome Med. 2010;2(12) doi: 10.1186/gm133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mei Y, Clark D, Mao L. Novel dimensions of piRNAs in cancer. Cancer Lett. 2013;336:46–52. doi: 10.1016/j.canlet.2013.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cheng J, Deng H, Xiao B, Zhou H, Zhou F, Shen Z, Guo J. piR-823, a novel non-coding small RNA, demonstrates in vitro and in vivo tumor suppressive activity in human gastric cancer cells. Cancer Lett. 2012;315:12–17. doi: 10.1016/j.canlet.2011.10.004. [DOI] [PubMed] [Google Scholar]
- 16.Cheng J, Guo JM, Xiao BX, Miao Y, Jiang Z, Zhou H, Li QN. piRNA, the new non-coding RNA, is aberrantly expressed in human cancer cells. Clin Chim Acta. 2011;412:1621–1625. doi: 10.1016/j.cca.2011.05.015. [DOI] [PubMed] [Google Scholar]
- 17.Huang G, Hu H, Xue X, Shen S, Gao E, Guo G, Shen X, Zhang X. Altered expression of piRNAs and their relation with clinicopathologic features of breast cancer. Clin Transl Oncol. 2013;15:563–568. doi: 10.1007/s12094-012-0966-0. [DOI] [PubMed] [Google Scholar]
- 18.Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL, Pachter L. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc. 2012;7:562–578. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Homann N, Tillonen J, Meurman JH, Rintamäki H, Lindqvist C, Rautio M, Jousimies-Somer H, Salaspuro M. Increased salivary acetaldehyde levels in heavy drinkers and smokers: A microbiological approach to oral cavity cancer. Carcinogenesis. 2000;21:663–668. doi: 10.1093/carcin/21.4.663. [DOI] [PubMed] [Google Scholar]
- 20.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 21.Kiang A, Yu MA, Ongkeko WM. Progress and pitfalls in the identification of cancer stem cell-targeting therapies in head and neck squamous cell carcinoma. Curr Med Chem. 2012;19:6056–6064. doi: 10.2174/0929867311209066056. [DOI] [PubMed] [Google Scholar]
- 22.Matta A, Ralhan R. Overview of current and future biologically based targeted therapies in head and neck squamous cell carcinoma. Head Neck Oncol. 2009;1:6. doi: 10.1186/1758-3284-1-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kleiber ML, Diehl EJ, Laufer BI, Mantha K, Chokroborty-Hoque A, Alberry B, Singh SM. Long-term genomic and epigenomic dysregulation as a consequence of prenatal alcohol exposure: A model for fetal alcohol spectrum disorders. Front Genet. 2014;5:161. doi: 10.3389/fgene.2014.00161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Maccani MA, Knopik VS. Cigarette smoke exposure-associated alterations to non-coding RNA. Front Genet. 2012;3:53. doi: 10.3389/fgene.2012.00053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Aalto AP, Pasquinelli AE. Small non-coding RNAs mount a silent revolution in gene expression. Curr Opin Cell Biol. 2012;24:333–340. doi: 10.1016/j.ceb.2012.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Siomi MC, Sato K, Pezic D, Aravin AA. PIWI-interacting small RNAs: The vanguard of genome defence. Nat Rev Mol Cell Biol. 2011;12:246–258. doi: 10.1038/nrm3089. [DOI] [PubMed] [Google Scholar]
- 27.Kim SK, Jung WH, Koo JS. Differential expression of enzymes associated with serine/glycine metabolism in different breast cancer subtypes. PLoS One. 2014;9:e101004. doi: 10.1371/journal.pone.0101004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim YH, Jung WH, Koo JS. Expression of metabolism-related proteins in invasive lobular carcinoma: Comparison to invasive ductal carcinoma. Tumour Biol. 2014;35:10381–10393. doi: 10.1007/s13277-014-2345-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets generated and/or analyzed during the current study are available in The Cancer Genome Atlas (https://portal.gdc.cancer.gov/projects/TCGA-HNSC).