Fig. 1.
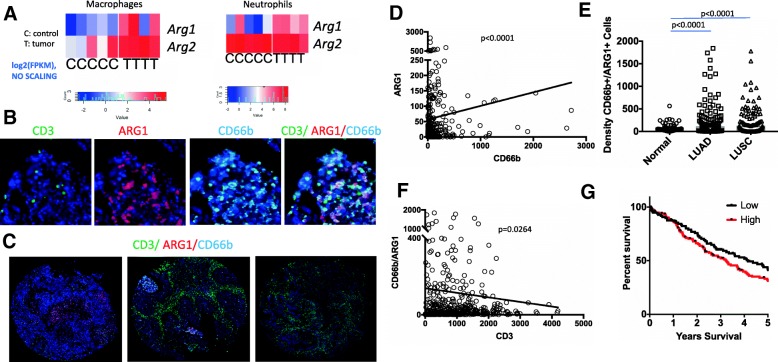
Arginase expression in the lung tumors. a RNA Sequencing analysis of sorted tumor (Epcam+), macrophage (CD11c+ CD11b- CD103-) and neutrophils (CD11b+, LY6G+) from KrasG12D lung tumors. T denotes cells isolated from tumor carrying mice and c from healthy control littermates. Heatmaps denote Log2 of fragments per kilobase of transcript (FPKM) per each mouse sample. p values are 0.02 and 0.03 for macrophages and neutrophils for Arg1 and 0.01 and 0.07 for macrophages and neutrophils for Arg2. Log2 (FPKM) values were also used for statistical analysis by student’s T test. Heatmaps represent unscaled values. b High power image of ARG1, CD3, CD66b multiplex immunohistochemistry. c Low power representative images from ARG1, CD3, CD66b multiplex immunohistochemistry staining of the TMA for varying levels of Arg1 and CD3 staining. Left: Arg1 high CD3 low, middle: Arg1 and CD3 high, and right: Arg1 low CD3 high tumor sample. d Correlation of CD66b and ARG1 in lung tumor samples. P < 0.0001, Spearman correlation. e Automated quantification of the ARG1 + CD66b + density in adjacent matched-normals, LUAD, and LUSQ p < 0.0001, determined by Mann-Whitney test f Correlation of CD66b+ ARG1+ double staining and CD3 staining in NSCLC, p < 0.0264, Spearman correlation. G. Kaplan Meier survival analysis for patients whose tumors were CD66b+/ARG1+ high or low density based on multiplex IHC. p = 0.09 (Log Rank test). The thresholds set for fluorescence positive signals for CD66b and CD3 are higher than that for ARG1 therefore the quantity of the expression is relative not exact