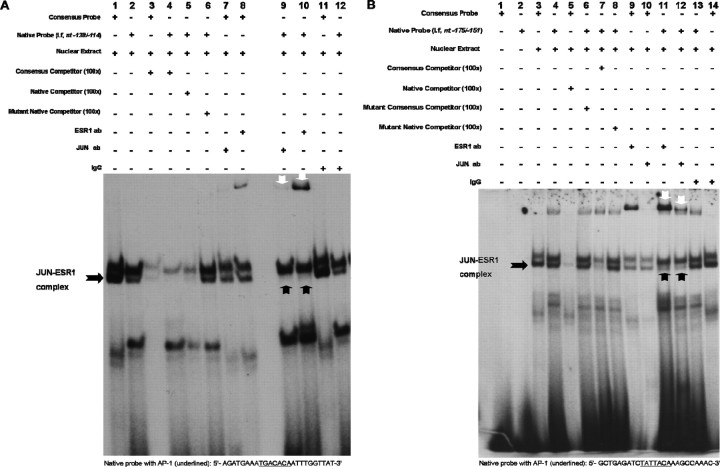
Fig. 6.
Putative AP-1 sites at nt −130/−124 and nt −166/−160 regions of promoter I.f are the major c-Jun-ESR1 binding sites. An EMSA was used to visualize JUN and ESR1 binding to AP-1 sites within promoter I.f. The EMSAs were performed using 5 μg of nuclear extract from N42 hypothalamic cells treated with E2 (10−7 M; 6 h) and radiolabeled oligonucleotide probes containing critical AP-1 sites and flanking sequences. A) Binding to an oligonucleotide identical to the nt −138/−114 sequence in promoter I.f and containing the nt −130/−124 AP-1 element (native; lane 2). Competition studies were performed using 100-fold excess unlabeled wild-type (consensus; lanes 3 and 4) or native oligonucleotides (lane 5) or mutant native oligonucleotides (lane 6). Immunodepletion of complexes was performed using 2 μg of anti-c-Jun (lanes 7 and 9) or 0.5 μg of anti-ESR1 (lanes 8 and 10) antibody (ab). Normal rabbit IgG was added as a negative control (lanes 11 and 12). B) Binding to an oligonucleotide identical to the nt −175/−151 sequence in promoter I.f and containing the nt −166/−160 AP-1 element (native) (lane 2). Competition studies were performed using 100-fold excess unlabeled consensus or native probes (lanes 5 and 7) or mutant consensus or mutant native probes (lanes 6 and 8). Immunodepletion of complexes was performed using 2 μg of anti-c-Jun (lanes 10 and 12) or 0.5 μg of anti-ESR1 (lanes 9 and 11) antibody (ab). Normal rabbit IgG was added as a negative control (lanes 13 and 14). Black arrows indicate JUN-ESR1 complexes, and white arrows indicate supershifted complexes by either c-Jun or ESR1 antibodies. The figures represent one of three independently performed experiments.