Fig. 2.
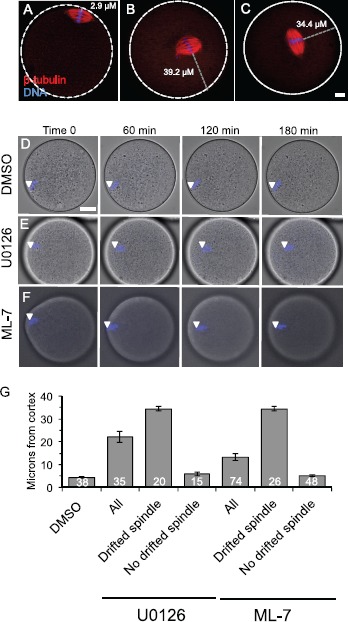
Spindle localization in U0126- and ML-7-treated metaphase II eggs. A–C) Fluorescence images show the localization of the metaphase II spindle in eggs stained with anti-β-tubulin (red) and DAPI to stain the maternal DNA (blue). White dotted lines show the perimeter of the egg. A shows a control egg with normal spindle localization. B and C show representative U0126- and ML-7-treated eggs in which the metaphase II spindle has drifted away from the cortex. The gray dotted line indicates how spindle distance was measured (in μm) using the point of the maternal DNA on the metaphase plate closest to the egg periphery as the start point, and the egg perimeter as the end point. Bar in C = 10 μm for A–C. D–F) Time-lapse microscopy of DAPI-loaded eggs over time, with eggs treated with DMSO (D), 50 μM U0126 (E), or 15 μM ML-7 (F). D and E show single optical sections (because the spindle was moving within this plane), and F is a three-dimensional maximum intensity projection (MIP) image from multiple optical sections. White arrowheads indicate the localization of the DAPI-stained maternal DNA. Bar in D = 17 μm for D–F. G) Average spindle-to-cortex distances (in μm) in DMSO-, U0126-, and ML-7-treated eggs. The U0126- and ML-7-treated eggs are further separated into the subgroup of eggs with drifted spindles (i.e., cortex-to-DNA distance greater than 12 μm) and the subgroup of eggs with no drifted spindle (i.e., cortex-to-DNA distance less than 12 μm). Control DMSO-treated eggs had spindles that were a maximum of 10.5 μm from the cortex (mean 5.2 ± 0.4 μm). See text for additional details.
