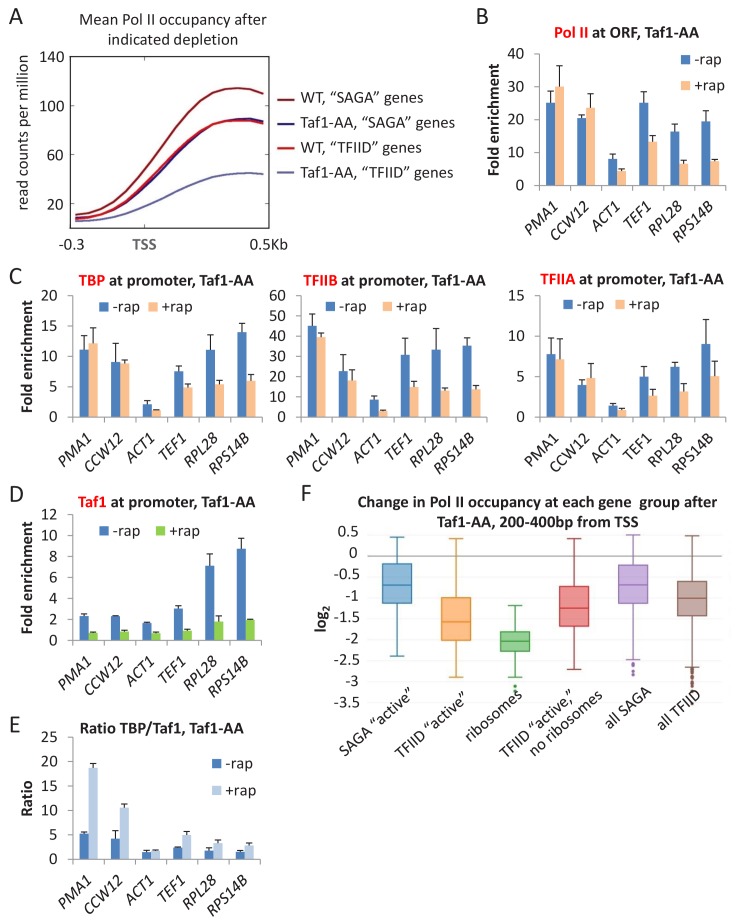
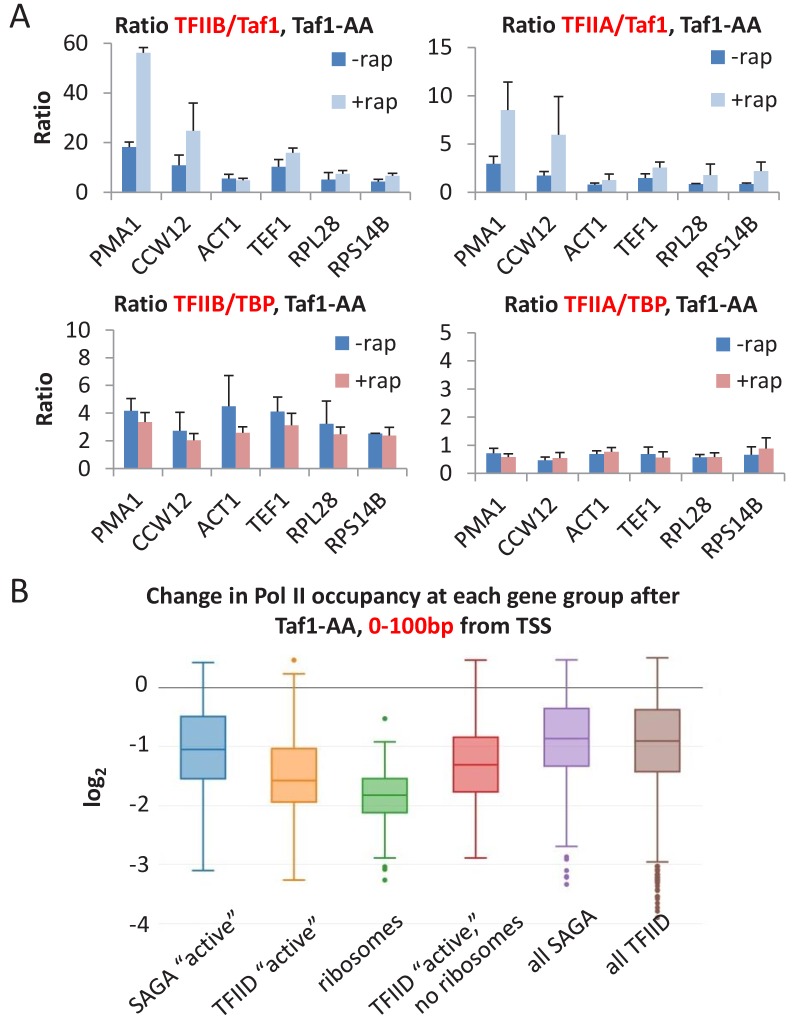
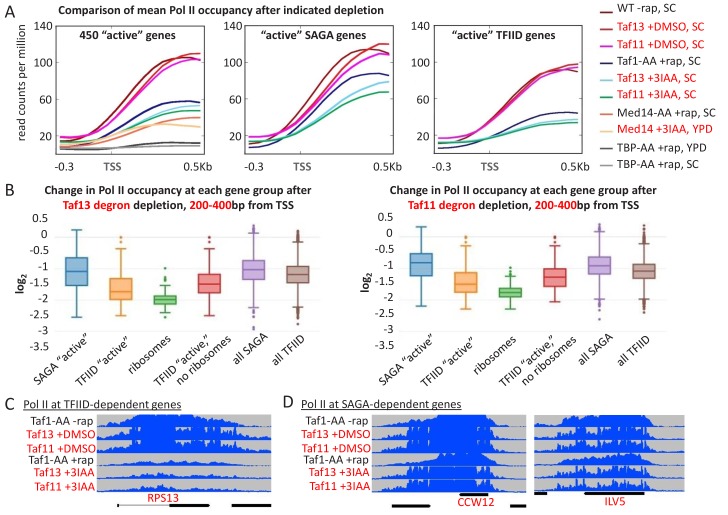
Figure 7. Taf1 depletion selectively affects TFIID-dependent genes.
(A) Mean Pol II occupancy averaged over 453 well-transcribed ‘SAGA’ or ‘TFIID’ genes in parental or Taf1-depleted strains. (B) Pol II (C) TBP, TFIIB, and TFIIA, and (D) Taf1 occupancies at ‘SAGA’ (PMA1 and CCW12) or ‘TFIID’ (ACT1, TEF1, RPL28, RPS14B) coding regions in the Taf1-depletion strain treated or untreated with rapamycin. (E) TBP:TAF1 occupancy ratios in the Taf1-depletion strain treated or untreated with rapamycin. (F) Log2 change in Pol II occupancy (measured at +200–400 from the TSS) for the indicated gene classes upon Taf1 depletion. ‘Active’ genes are the top 10% of transcribed genes, broken up into ‘SAGA’ and ‘TFIID’-dependent categories. Ribosomes include only the ribosomal genes from the TFIID-dependent category. ‘TFIID active, no ribosomes’ are the remainder of the top 10% transcribed genes in the TFIID-dependent category after ribosomal genes have been removed. ‘All’ genes are the entire set of ~5000 s. cerevisiae genes, broken up into ‘SAGA’ and ‘TFIID’-dependent categories.