Figure 2. Single cell proliferation rate distributions for 1500 gene deletions.
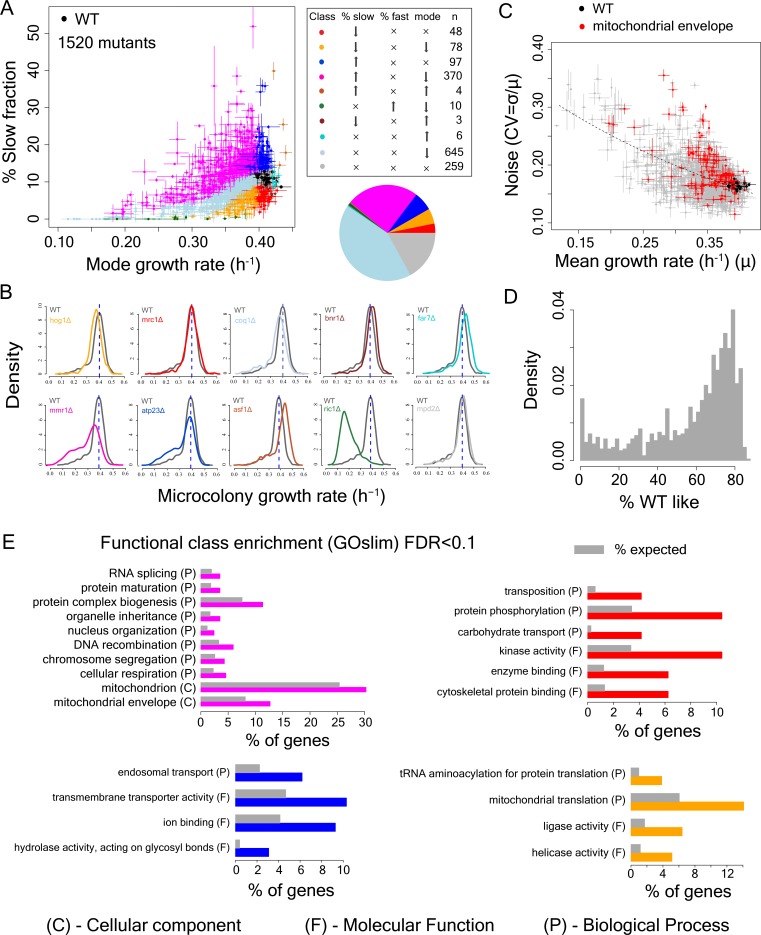
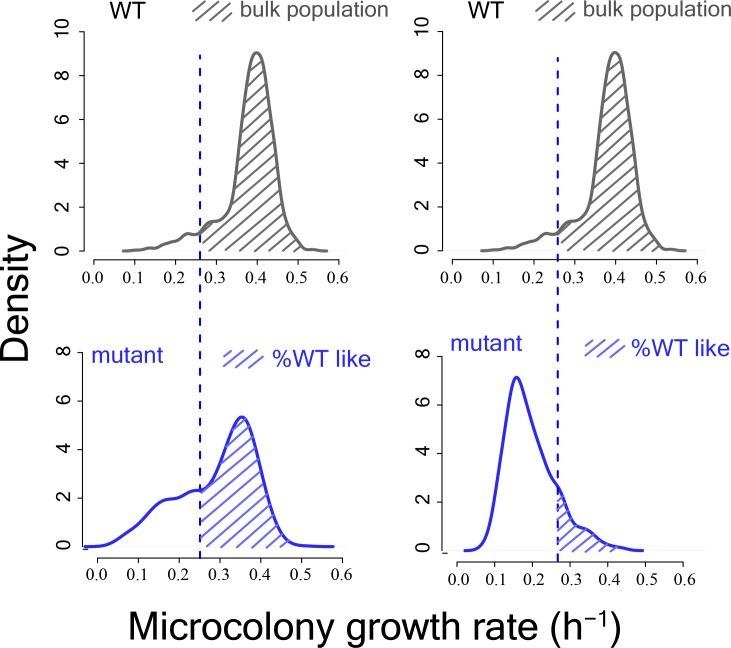
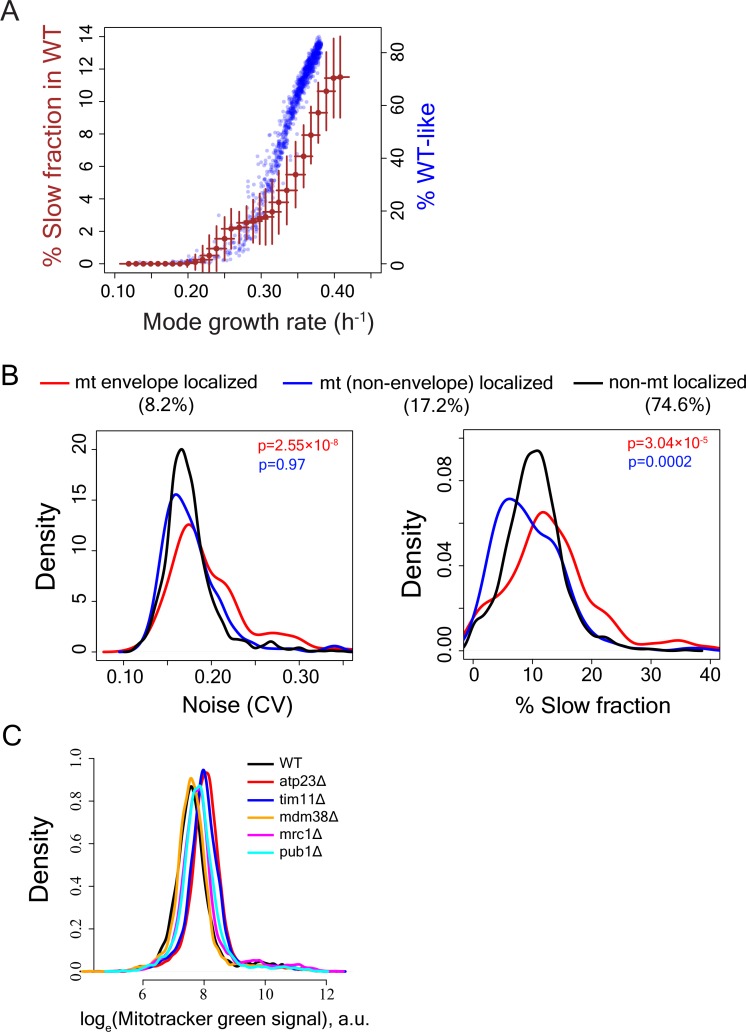
(A) Mode growth rate (h−1) and % slow fraction for 1520 deletion strains. The points represent average values across replicates and the bars represent ±1 s.d. values. The colours show classification of mutants into different categories according to change in mode growth rate (see Materials and methods, FDR < 0.1) and change in % slow fraction (FDR < 0.1) compared to the wild-type (WT) strain. The table and pie chart show the number and proportion of strains in each group (colour coded). Replicate data for WT strain are shown by multiple black points. (B) Examples of growth distributions of mutants classified into different groups which are colour coded as in A. The distribution in dark grey shows WT growth distribution. (C) Coefficient of variation (CV) vs. mean growth rate for all strains. WT values are shown in black; mutants of genes that localize to mitochondrial envelope in red. The points represent average values across replicates and the bars represent ±1 s.d. values. (D) % of WT-like cells in all mutants showing variable mutation outcome. It was calculated for all mutants showing significant reduction in mean proliferation rate and had significant proportion of cells growing as fast as the bulk of the WT proliferation distribution (Wilcoxon rank-sum test). (E) Functional class enrichment (GOslim) analysis for different classification groups show significantly enriched functional classes (hypergeometric test, FDR < 0.1). P – Biological Process, F – Molecular Function, C- Cellular Component. Bars show % of genes in a particular group (colour coded) being present in that particular functional class.