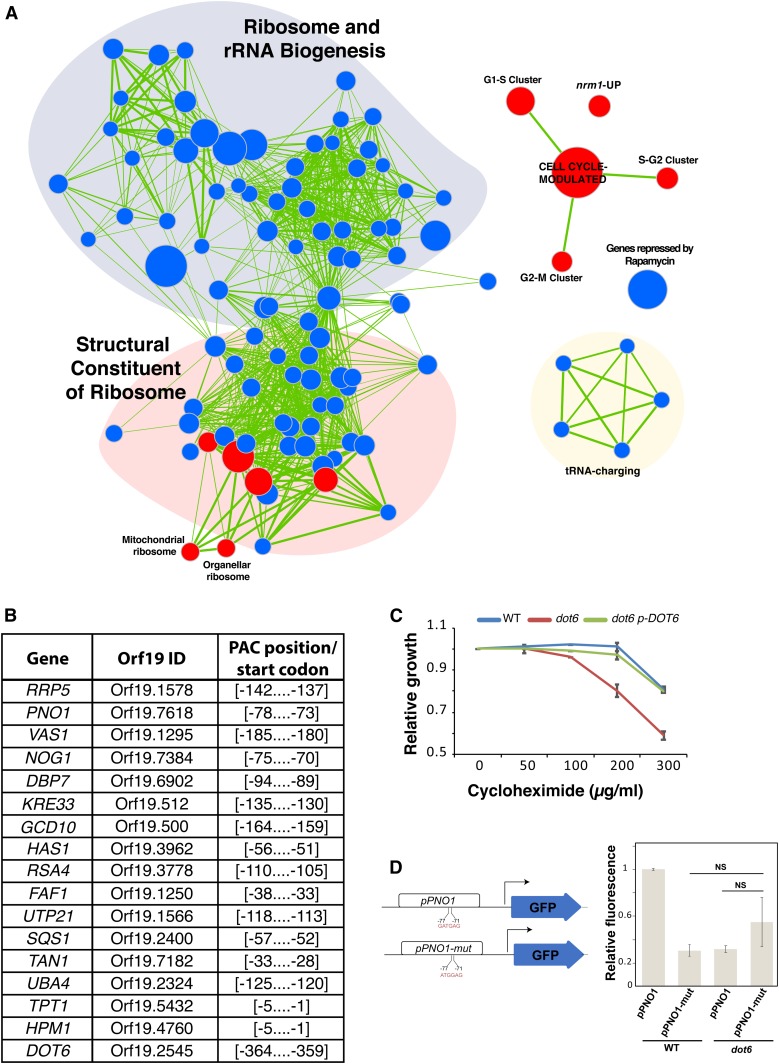
Figure 3.
Dot6 is a positive regulator of ribosome biogenesis genes. (A) GSEA analysis of differentially expressed genes in a dot6 mutant relative to the WT strain (SFY87). Cells were synchronized in G1 phase by centrifugal elutriation, released in fresh SC medium for 10 min, and analyzed for gene expression profiles by DNA microarrays. Correlations of dot6 upregulated (red circles) and downregulated (blue circles) transcripts are shown for biological processes, gene lists in different C. albicans mutants and experiments. The diameter of the circle reflects the number of modulated gene transcripts in each gene set. Known functional connections between related processes are indicated (green lines). Images were generated in Cytoscape with the Enrichment Map plug-in. (B) Occurrence of the PAC motif in the promoters of Dot6-modulated Ribi genes. The 400 bp sequence upstream the start codon of downregulated genes in dot6 (transcript with 1.5-fold reduction using 5% FDR) were scanned for the GATGAG motif. (C) Effect of the translation inhibitor cycloheximide on the growth of the WT (SFY87), dot6 mutant, and the revertant (dot6 p-DOT6) strains. Strains were grown on in SC medium at 30° for 24 hr. Relative growth was calculated as fraction of OD600 of cycloheximide-treated cells relatively to the nontreated controls. Results are the mean of three replicates. (D) GFP reporter assay to confirm that the transcription at the PNO1 (Orf19.7618) locus is driven by the Dot6 PAC-binding element. The pPNO1-GFP reporter strain was constructed by replacing one copy of the PNO1 ORF by the GFP ORF. Mutation in the PAC motif of the pPNO1-mut strain was introduced by PCR using the forward primer pPNO1mut-GFP-F. GFP fluorescence was measured by flow cytometry, and results are presented as relative mean GFP fluorescence as compared to pPNO1-GFP construct in the WT strain. Bars show the means ± SEM. NS, not significant (P > 0.15).