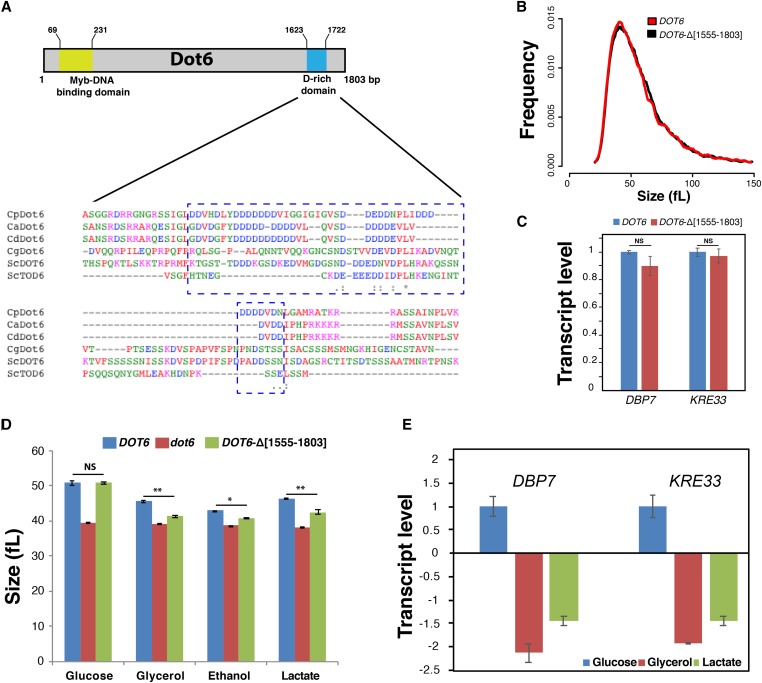
Figure 5.
The CTG-clade specific acidic domain of Dot6 is required for size control in response to nonfermentable carbon sources. (A) The C-terminal D-rich domain of Dot6 is conserved in the CTG clade species C. albicans (Ca), C. parapsilosis (Cp) and C. dubliniensis (Cd), but not in S. cerevisiae (Sc) and C. glabrata (Cg). Identical residues are indicated with asterisks. Conserved and semiconserved substitutions are denoted by colons and periods, respectively. (B) Cell size distribution of the WT (SC5314) and the truncated DOT6-Δ[1555–1803] strains. (C) Transcript levels of Ribi genes, including DBP7 and KRE33, were evaluated in both WT (SC5314) and the truncated DOT6-Δ[1555–1803] strains. Transcript levels were calculated using the comparative CT method using the ACT1 gene as a reference. Results are the mean of three replicates. For each transcript, fold changes in the WT and the truncated strains were not statistically significant (t-test). NS, not significant. (D) Median size of the WT (SC5314), dot6, and the truncated DOT6-Δ[1555–1803] strains growing in YP-glucose, YP-glycerol, YP-ethanol, and YP-lactate media. Results are the mean of three independent replicates. * P < 0.02; ** P < 0.01; (E) Transcript levels of Ribi genes, DBP7, and KRE33 in both WT (SC5314) and the truncated DOT6-Δ[1555–1803] strains growing in fermentable (glucose) and nonfermentable carbon sources (glycerol and lactate). Values represent transcript levels of DBP7 and KRE33 in cells growing in glycerol or lactate normalized to that of cells growing in glucose. Results are the mean of three replicates.