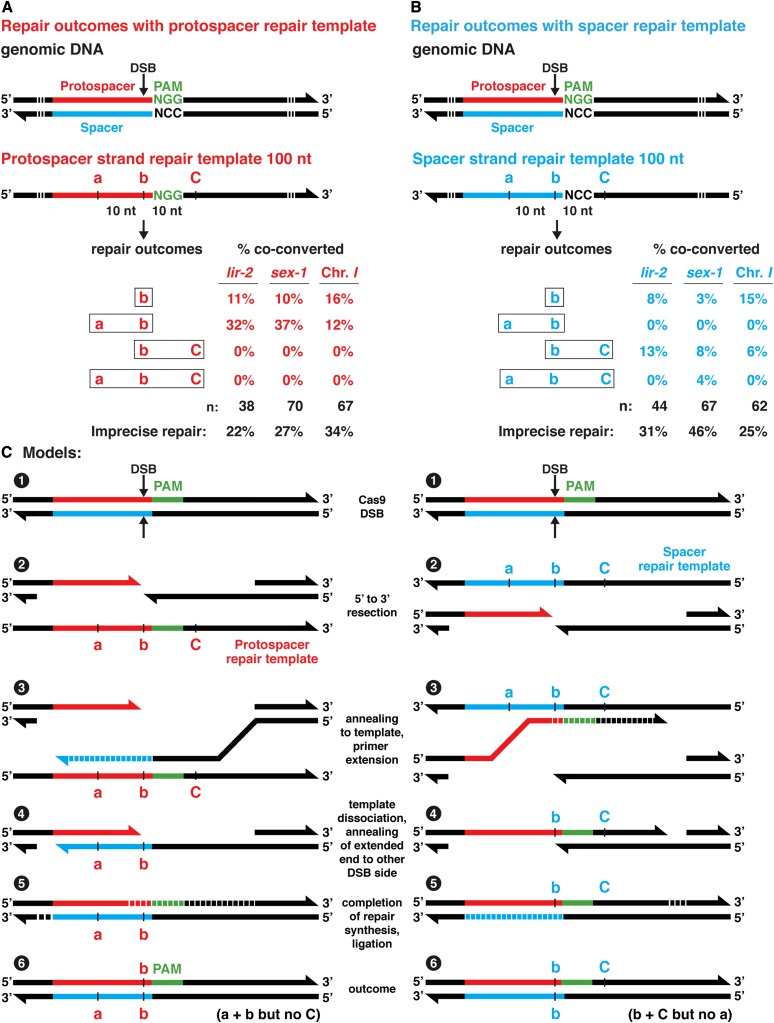
Figure 2.
Polarity of HDR from single-stranded templates is dictated by the choice of repair template strand. (A and B) Analysis of genome editing experiments at three loci (lir-2, sex-1, intergenic chromosome I site) comparing HDR outcomes using Cas9 RNPs and single-stranded repair templates of either (A) the protospacer strand or (B) spacer strand. Polymorphisms were located in the repair templates at the predicted endogenous DSB sites (SNP b) and at sites both 5′ (SNP a) and 3′ (SNP C) of the PAM (Table S2). Percentages represent the frequencies of diagrammed repair outcomes at each locus relative to the total number of mutants (n) expressing the co-conversion marker in that experiment. Both protospacer and spacer repair templates promoted insertion of SNPs at the DSB site, but the protospacer strand template promoted insertion of SNPs exclusively 5′ of the PAM, while the spacer strand template promoted insertion of SNPs preferentially 3′ of the PAM. Frequency of imprecise repair is also provided for each experiment. For these experiments, dpy-10(gf) was the co-conversion marker. When precise templated HDR occurred at the dpy-10 locus, the animals exhibited a dominant Rol phenotype and a recessive Dpy phenotype. If imprecise repair occurred at dpy-10, animals exhibited only a recessive Dpy phenotype. All animals with any of these phenotypes were examined for SNPs. (C) Models show proposed steps in the HDR process for protospacer (left) and spacer (right) strand repair templates based on a repair mechanism involving SDSA. Numbers refer to sequential steps.