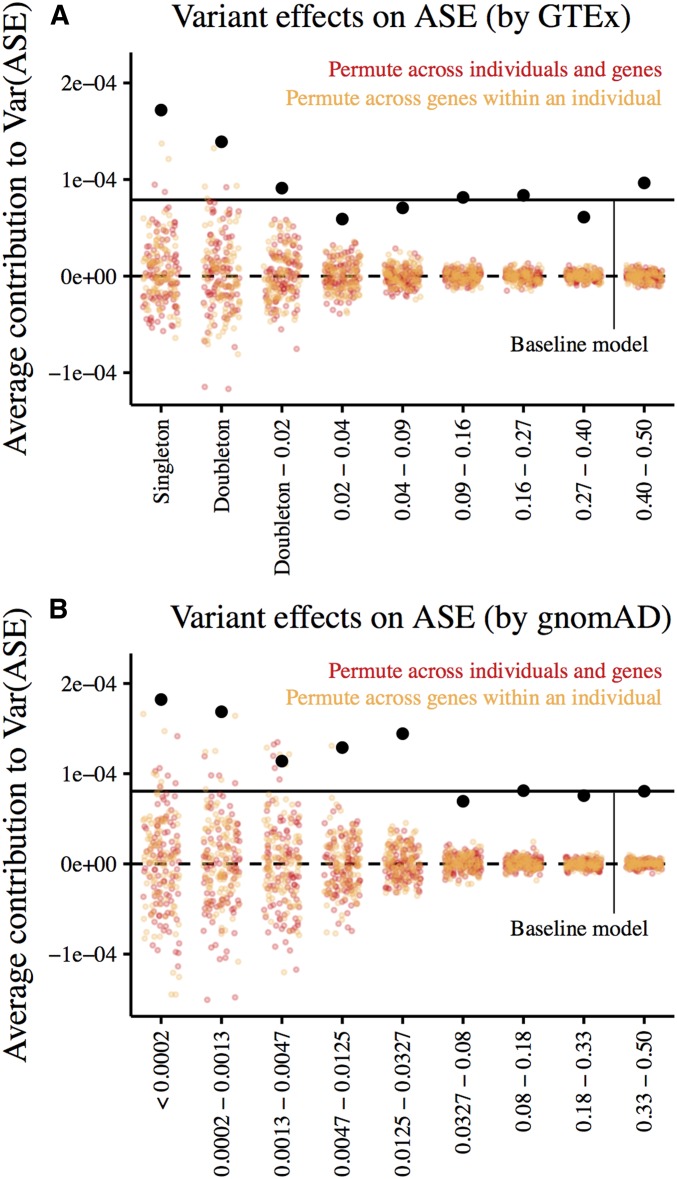
Figure 7.
Effects of cis-genetic variants on allelic imbalance given their allele frequencies. Estimates are obtained from multiple regression on allelic imbalance using the number of cis-heterozygous sites in each allele frequency bin as predictors. Here, we include sites in a 20-kb window centered on the TSS of a gene with measured allelic imbalance. Estimates from combined-tissue ASE (reads spanning each ASE site in a single individual are summed across all tissues sampled) are shown in black. Colored points represent estimates from 100 permutations of (1) ASE measurements across genes and individuals (red) and (2) ASE measurements across genes within an individual (orange). Horizontal lines mark zero effect (dashed) and the average variant effect estimated by regressing allelic imbalance on the total number of cis-heterozygous sites (solid). (A) Estimates with variants binned by minor allele frequency in Europeans in the GTEx data (n = 122). (B) Estimates with variants binned by minor allele frequency in Europeans in gnomAD (n = 7509).