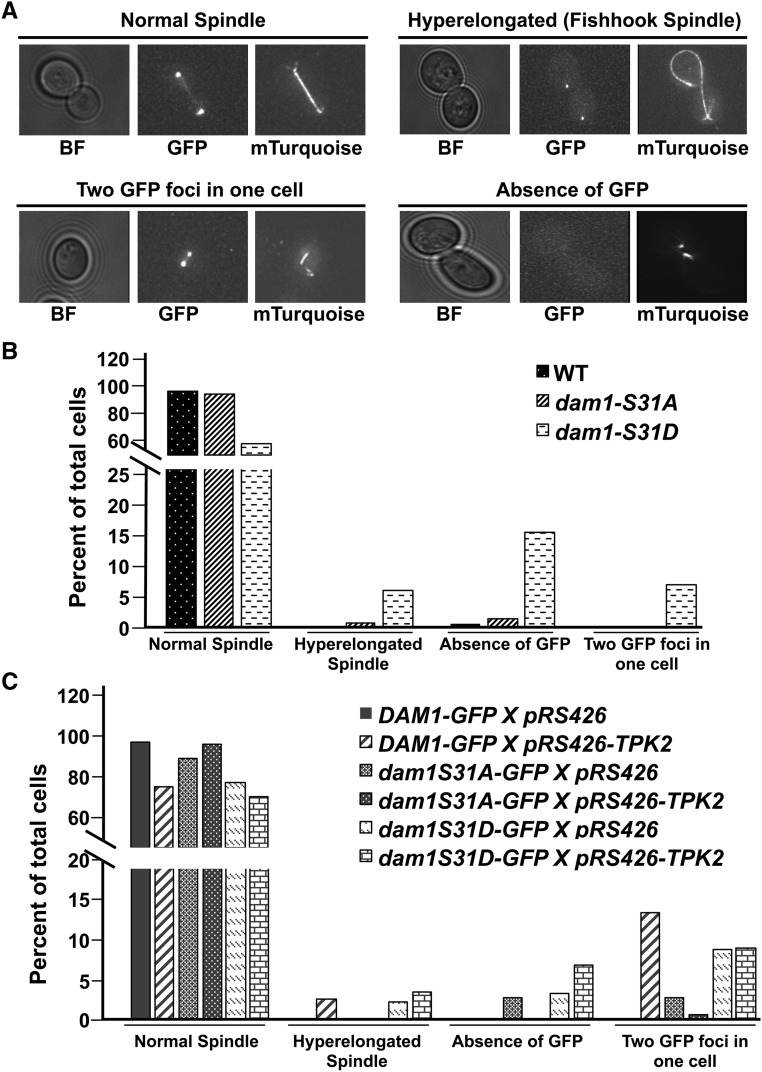
Figure 5.
Abnormal spindle morphology and chromosome segregation defects in dam1p-S31D phospho-mimetic mutants and in wild-type cells carrying high-copy TPK2. (A) Shown are representative images for four different phenotypes: “normal spindle,” “hyper-elongated spindle” (fishhook spindle), “two GFP foci” (refers to two GFP foci in one unbudded cell or in one cell of a large budded cell), and “absence of GFP” (refers to an absence of GFP in unbudded or large budded cells). All GFP and mTurquoise images shown are the maximum intensity projections of Z-sections of the stacks onto a single plane. Wild-type Dam1p-GFP, Tub1p-Turquoise (JCY1036); dam1p-S31A-GFP, Tub1p-Turquoise (JCY1036); and dam1p-S31D-GFP, Tub1p-Turquoise (JCY1037) strains were grown in SC media as described in Materials and Methods and imaged. Images that represent normal spindle are taken from Dam1p-GFP, Tub1p-Turquoise; hyper-elongated spindle is from dam1p-S31D-GFP, Tub1p-Turquoise; unbudded cell with two GFP foci is taken from Dam1p-GFP carrying high-copy TPK2; and absence of GFP is taken from dam1p-S31D-GFP. (B) Percent of total cells in four different categories were plotted. Number of cells counted are: Dam1p-GFP, n = 216; dam1p-S31A-GFP, n = 143; and dam1p-S31D-GFP, n = 193. (C) Wild-type Dam1p-GFP, Tub1p-Turquoise (JCY1036); dam1p-S31A-GFP, Tub1p-Turquoise (JCY1036); and dam1p-S31D-GFP, Tub1p-Turquoise (JCY1037) strains carrying 2 µm empty vector, pRS426 (JCB63), or high-copy TPK2, pRS426-TPK2 (JCB59) were grown in SC-URA as described in Materials and Methods and imaged. Percent of total cells are plotted. Numbers of cells counted are:. Dam1p-GFP pRS426, n = 121; Dam1p-GFP pRS426-TPK2, n = 76; dam1p-S31A-GFP pRS426, n = 69; dam1p-S31A-GFP pRS426-TPK2, n = 155; dam1p-S31D-GFP pRS426, n = 175; and dam1p-S31D-GFP pRS426-TPK2, n = 138. BF, bright field; GFP, Dam1p-GFP (and mutants); mTurquoise, Tub1p-mTurquoise.