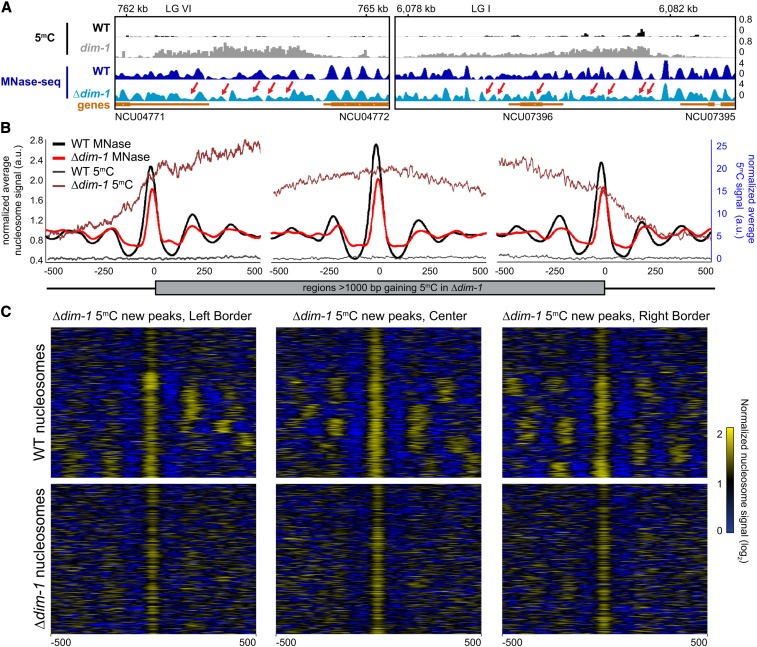
Figure 5.
Intergenic regions that gain cytosine methylation in Δdim-1 exhibit aberrant nucleosome positioning. (A) Representative examples of nucleosome positions within intergenic regions that gain cytosine methylation. Tracks of bisulfite-sequencing for WT (black) and dim-1 (light gray), as well as MNase-sequencing for WT (dark blue) and Δdim-1 (light blue) strains are shown. Red arrows highlight changes in nucleosome positioning in Δdim-1 strain. (B) Average mono-nucleosome enrichment profiles (MNase) and cytosine methylation (5mC) in WT and Δdim-1 strains at the first nucleosome apex in left and right borders of regions that gain methylation in a dim-1 strain, as well as a central reference nucleosome peak in a WT strain (n = 322); the WT data used reference nucleosomes in a WT strain while the dim-1 data used reference nucleosomes from a Δdim-1 strain. (C) Heatmaps of nucleosome signals from WT and Δdim-1 strains normalized to average nucleosome signal examining ±500 bp of the intergenic regions gaining cytosine methylation in Δdim-1 strain shown in B; regions are shown in each heatmap pair in the same order.