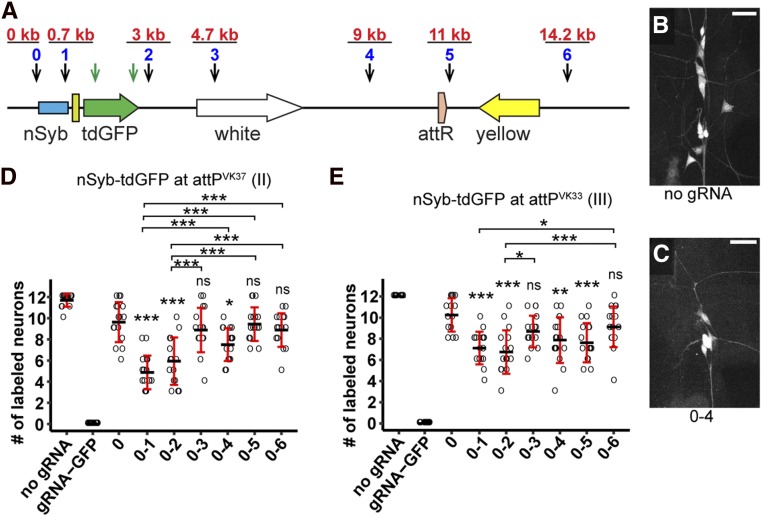
Figure 3.
Efficiency of dual guide RNA (gRNA)-mediated DNA deletion at the single-cell level. (A) Diagram showing the nSyb-tdGFP reporter integrated in the genome and gRNA target sites. Each blue number and the black arrow below it indicate a gRNA-targeting noncoding sequence of the reporter. The distance of each gRNA from gRNA 0 is indicated in red above the gRNA. The two green arrows indicate two gRNAs targeting the coding sequence of tdGFP. (B and C) Dorsal clusters of PNS neurons labeled by the reporter in a control animal (B) and an animal expressing gRNAs 0 and 4 (C). Bar, 25 μm. (D and E) Quantification of the number of GFP-positive neurons for each gRNA pair using nSyb-tdGFP inserted at attPVK37 (D) and attPVK33 (E) sites. Each circle represents an individual neuron (n = 16 neurons for each genotype). Black bar, mean; red bars, SD. * P ≤ 0.05, *** P ≤ 0.001; one-way ANOVA and Tukey’s honest significant difference test.