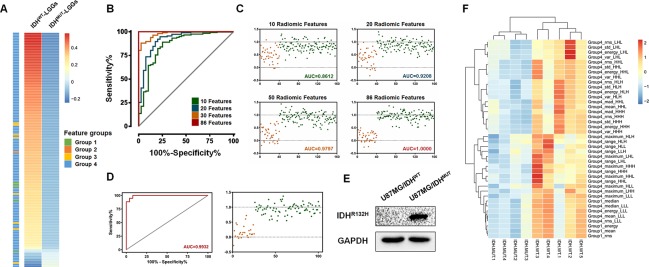
Figure 2.
Identification and validation of the IDH mutation-specific radiomic signature using the logistic regression. (A) A total of 146 radiomic features were selected using SAM methods. The mean value and the corresponding groups of the differentially expressed features are listed. (B and C) In the training set, the logistic regression-derived radiomic features was able to separate LGGs into two groups with high sensitivity and specificity. The AUCs were 0.86, 0.92, 0.98 and 1.00 for 10, 20, 50 and 86 radiomic features, respectively. (D) Importantly, these 86 features comprised a signature enabling the distinction of LGGs into IDHMUT and IDHWT groups with an AUC of 0.9932. (E) A Western Blot assay confirmed the expression of the mutant IDH1 protein (IDH1R132H, 1:200, DIA-H05, Dianova). (F) The radiogenomic analysis of xenograft gliomas of nude mice. Differential radiomic features between LGGs patients could be used to distinguish the IDH mutation phenotype in the xenograft model as well.