Figure 6.
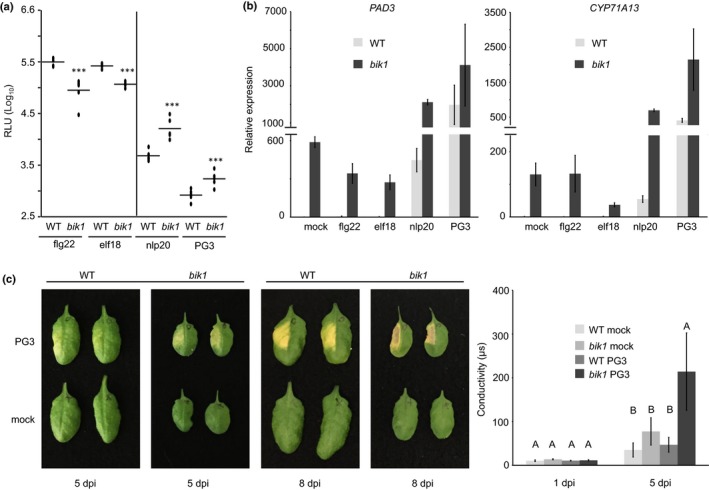
BIK1 plays a differential role in LRR‐RP and LRR‐RK signaling. (a) Arabidopsis leaf discs of wild‐type (WT) or bik1 mutant plants were treated with each 0.5 μM flg22, elf18, nlp20, or 0.1 μM PG3 and reactive oxygen species (ROS) accumulation (relative fluorescence units, RLU) was determined. Peak value minus background value are shown as dots (n ≥ 6) and means are presented as lines. Asterisks indicate significant differences compared with the wild‐type control treatment as determined by Student's t‐test: ***, P < 0.001. (b) Transcriptional profiling of camalexin biosynthesis genes by quantitative real‐time PCR (qRT‐PCR). Leaves of Arabidopsis wild‐type or bik1 mutant plants were infiltrated with water (mock), each 0.5 μM flg22, elf18, nlp20, or PG3 and collected 6 h after treatment. Relative expression of the indicated genes was normalized to the levels of the EF‐1α transcript and calibrated to the levels of the mock treatment in the WT control. Data present means of three technical replicates ± SD. (c) Leaves of Arabidopsis WT or bik1 mutant plants were infiltrated with water (mock) or 0.5 μM PG3 and visible cell death symptoms were monitored at 5 and 8 d post infiltration (dpi) (left panel). In addition, cell death progression 1 and 5 dpi was monitored by ion leakage assays using excised leaf discs floating on water (right panel). Bars represent mean conductivity ± SD (n = 4), capital letters indicate significant differences (P < 0.001) as determined by Student's t‐test. Each experiment was performed three times with similar results.
