Fig. 1.
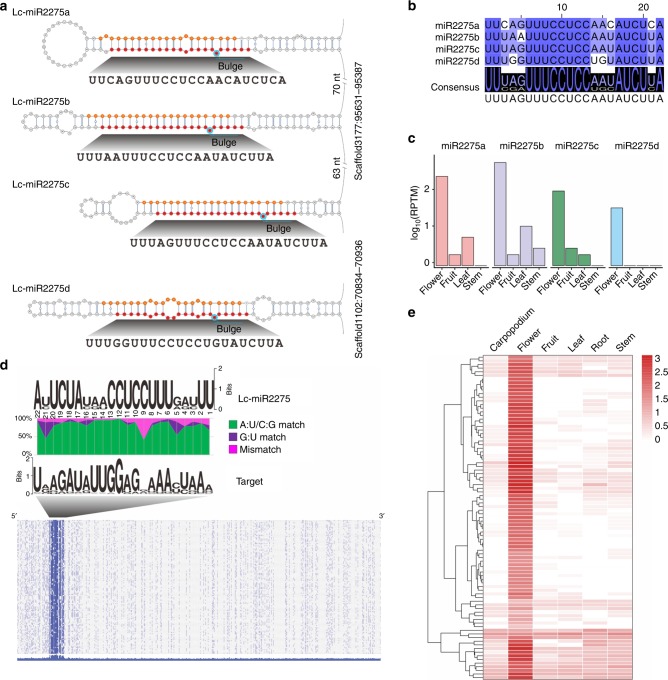
miR2275-triggered 24-nt reproductive phasiRNAs are abundant in litchi. a Stem–loop regions of two MIR2275 precursors in litchi. miR2275 and miR2275* sequences are marked in red and orange, respectively. Bulges within the miR2275/miR2275* duplexes are indicated by a cyan circle. b Alignment of all expressed members of the miR2275 family in litchi. The degree of conservation for each nucleotide is represented by the color, with a dark color denoting a high level of conservation and a light color denoting a low level. The consensus sequence of the alignment is displayed below with sequence logos. c Expression of miR2275 members in four litchi tissues. d Above: alignment of miR2275 duplex and its target sites in 24-PHAS loci in litchi. The nucleotide pairing at each position between miR2275 and miR2275* is indicated by different colors, with A:U/C:G matches denoted in green, G:U matches in purple, and all mismatches in pink. Below: nucleotide sequence alignment of 24-PHAS loci with sequence similarity denoted by the density of blue color, demonstrating no conservation outside of the miR2275 target site (as in grass 24-PHAS loci). e Accumulation of 24-nt phasiRNAs in different tissues of litchi, with each row denoting a 24-PHAS locus, and each column a tissue as indicated. The key at right indicates the abundance in units of log2(RPTM)