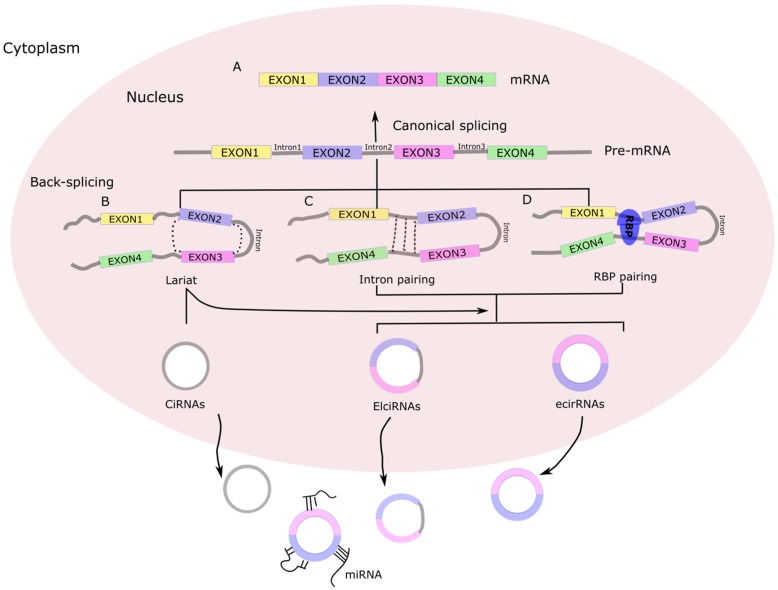
Figure 1.
Biogenesis of circRNA. (A) Canonical splicing to form mRNA. (B) Lariat-driven circularization. First, a pre-mRNA is spliced, causing the 3'-hydroxyl of the upstream exon to covalently bind to the 5'-phosphate of the downstream exon. At the same time, the sequence between the exons becomes an RNA lariat, containing several exons and introns. Second, in the RNA lariat, the 2'-hydroxyl of the 5'-intron reacts with the 5'-phosphate of the 3'-intron, followed by the 3'-hydroxyl of the 3'-exon reacting with the 5'-phosphate of the 5'-exon. As a result, an RNA double lariat and a circular RNA are produced. Finally, some introns of the circular RNA are removed, producing an ecirRNA, EIciRNA, or ciRNA. (C) Intron pairing-driven circularization. The circular structure can be generated through direct base-pairing of the introns flanking inverted repeats or complementary sequences. The introns are removed or retained to form ecirRNA or EIciRNA. (D) RNA binding proteins (RBPs)-driven circularization. In this case, RBPs bind the upstream and downstream introns. The RBPs are attracted to each other, and form a bridge between the introns. The 2'-hydroxyl of the upstream intron then reacts with the 5'-phosphate of the downstream intron, which is followed by the 3'-hydroxyl of the 3'-exon reacting with the 5'-phosphate of the 5'-exon. Some introns of the circRNA are ultimately removed, producing an ecircRNA or EIcirRNA. CiRNAs, intronic circRNAs; ecirRNAs, exonic circRNAs; EIciRNAs, exon-intron circRNAs.