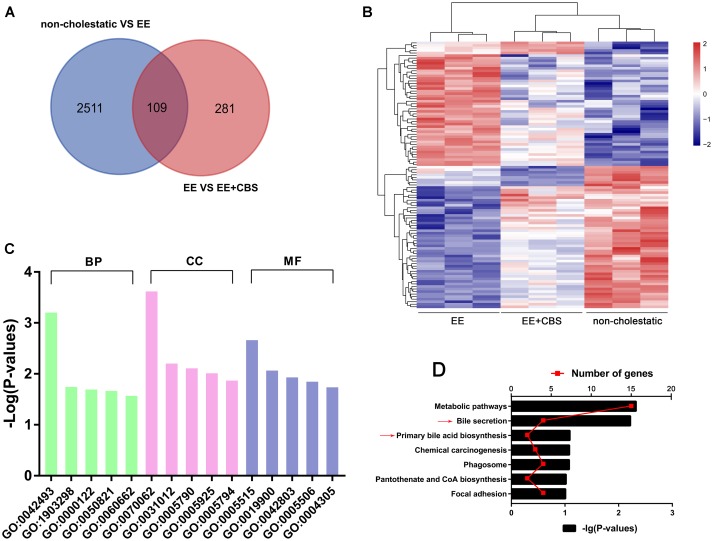
FIGURE 3.
DNA microarray analysis of non-cholestatic, 17α-ethinylestradiol (EE) and EE+CBS rat livers. (A) Venn diagram analysis of differentially expressed genes of non-cholestatic, EE and EE+CBS groups. (B) Heat map of differentially expressed genes in non-cholestatic, EE and EE+CBS groups (C) gene ontology (GO) analysis of differentially expressed genes (D) Kyoto encyclopedia of genes and genomes (KEGG) pathway analysis of differentially expressed genes. Arrows point to the pathways related to bile acid (BA) homeostasis; (n = 3). Abbreviations: BP, biological process; CC, cellular component; MF, molecular function. Interpretation: GO:0042493, response to drug; GO:1903298, negative regulation of hypoxia-induced intrinsic apoptotic signaling pathway; GO:0000122, negative regulation of transcription from RNA polymerase II promoter; GO:0050821, protein stabilization; GO:0060662, salivary gland cavitation; GO:0070062, extracellular exosome; GO:0031012, extracellular matrix; GO:0005790, smooth endoplasmic reticulum; GO:0005925, focal adhesion; GO:0005794, Golgi apparatus; GO:0005515, protein binding; GO:0019900, kinase binding; GO:0042803, protein homodimerization activity; GO:0005506, iron ion binding; and GO:0004305, ethanolamine kinase activity.