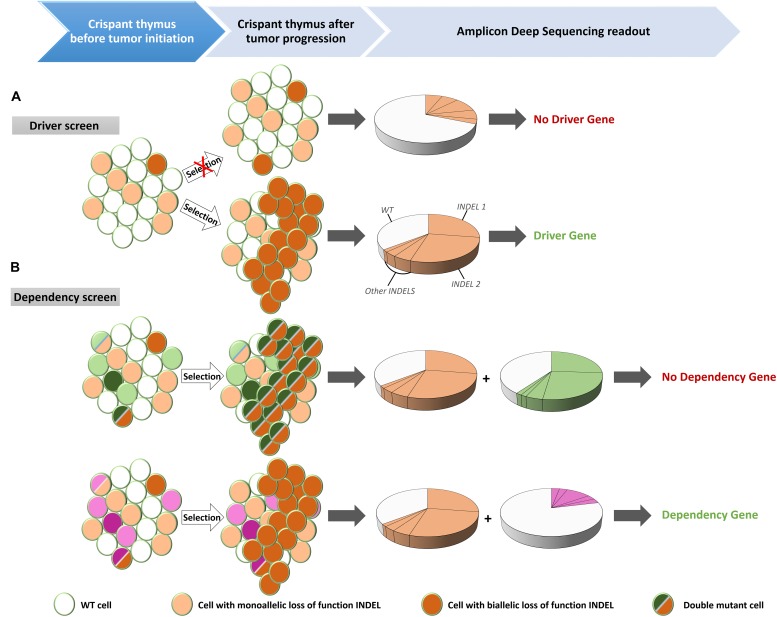
FIGURE 2.
Experimental set-up for the identification of driver mutations in tumor suppressor genes (TSG) and for dependency factors. (A) Injection of Cas9 recombinant proteins and guide RNAs targeting putative TSGs created mosaic mutant animals (crispants) that contain cells with mono-allelic or bi-allelic inactivation of the driver genes. Cells with bi-allelic inactivation will have a proliferative advantages over the wild type (WT) or mono-allelic mutant cells. When disease is observed, DNA is extracted from the thymus and after PCR amplification of the targeted regions, the PCR amplicon is subjected to deep sequencing. The presence of two unique frame shifting INDELs at equal proportion is indicative of a clonally expanding subpopulation, characteristic for leukemic disease. (B) Multiplexing of guide RNAs targeting a known driver gene together with a candidate dependency factor gene. If the candidate is not a dependency factor, clonal expanding cells will be present – identified by the INDEL signature of the driver gene (orange) – with bi-allelic inactivation of the analyzed gene (green) (top). However, in case of a genuine dependency factor gene (purple), expanding clones will never contain bi-allelic inactivation of this gene (bottom).