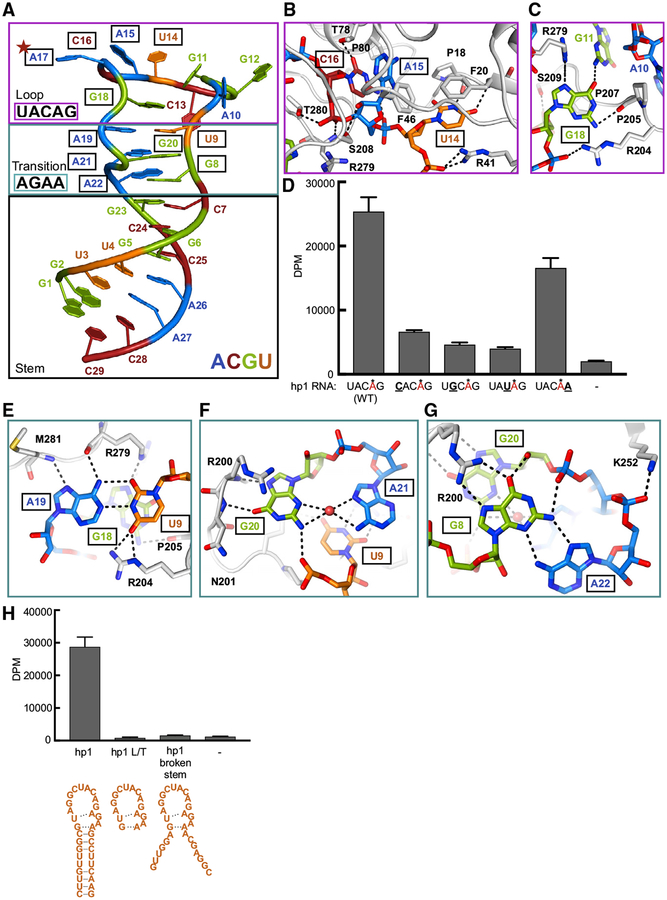
Figure 2. Specific RNA Loop Sequence Required for METTL16 Activity.
(A) Overall conformation of hp1x in the complex structure with MTD16. Nucleotides are numbered from 5′ to 3′. Target adenosine for methylation is indicated with a red star. Nucleotides in the conserved consensus motif of METTL16 have boxed labels.
(B and C) Detailed interactions in the RNA loop region. RNA nucleotides are shown as sticks, color-coded by base identity as in (A). Nucleotides within the consensus sequence are outlined with boxes: UAC (14–16) (B) and G18 (C). Protein residues are shown as gray cartoon and side-chain sticks, labeled with black text. Hydrogen bonds are marked with dashed lines.
(D) In vitro methylation activity of METTL16 on wild-type hp1 and hp1 with the indicated loop mutations. Methylated Ade is in red with an asterisk. Mutated base is boldfaced and underlined. Bars correspond to amounts of tritium incorporated into methylated RNA substrates shown as disintegrations per minute (DPM). Data are shown as mean DPM ± SD from three replicates.
(E–G) Detailed interactions in the transition region of hp1x. Each panel contains a pair of bases, where (E) contains the pair closest to the loop region, (F) is the next layer of the transition region, and (G) is the final pair, closest to the stem region. RNA bases are shown as sticks, color-coded as in (A). Nucleotides within the consensus sequence are outlined with black boxes. Polypeptide is shown as gray cartoon, with side chains represented with sticks. The ordered water molecule is shown as a red sphere. Hydrogen bonds are shown as dashed lines.
(H) In vitro methylation activity of METTL16 on the indicated RNA substrates shown as schematics underneath. Data are shown as mean DPM ± SD from three replicates.
See also Figure S2.