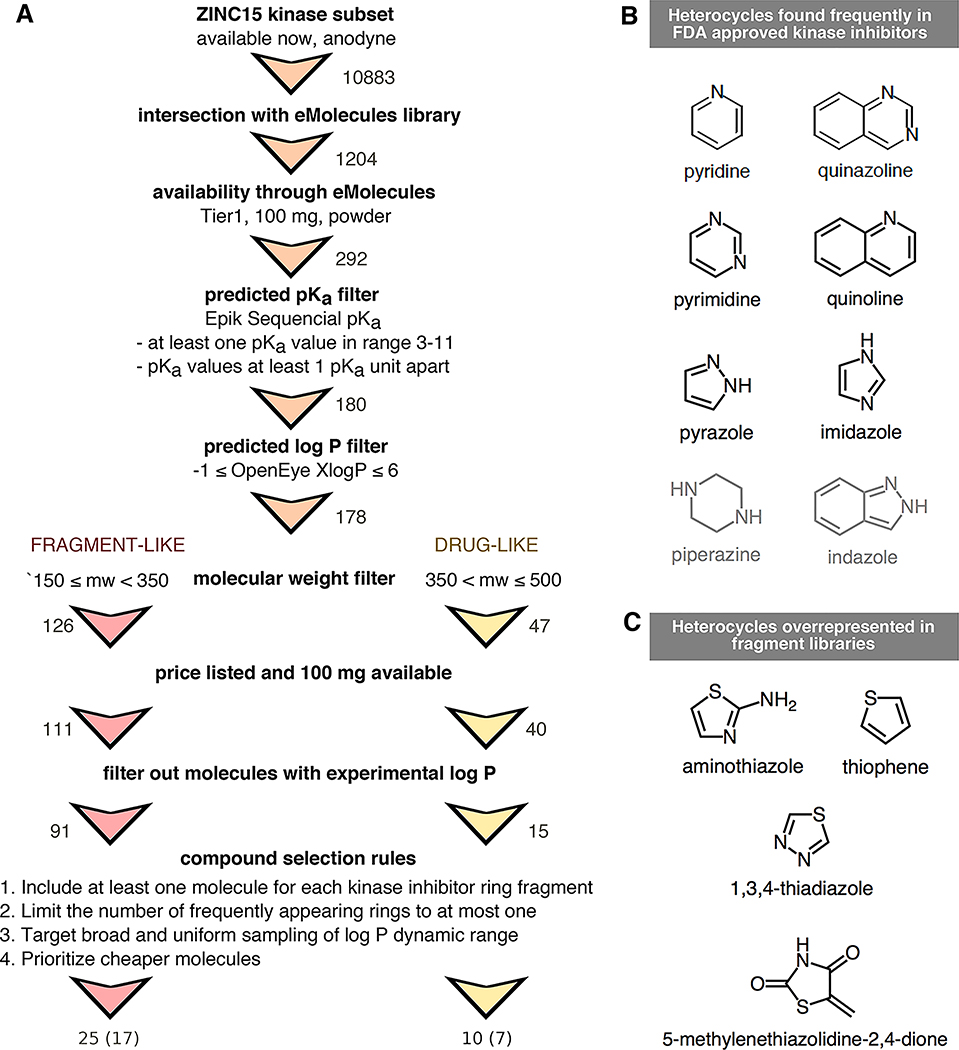
Figure 4. Compound selection for the SAMPL6 pKa challenge, with the goal of running subsequent log P/log D challenges on the same compound set.
(A) Flowchart of filtering steps for the selection of compounds that resemble kinase inhibitors and their fragments. Numbers next to arrows indicate the number of compounds remaining after each filtering step. A total of 25 fragment-like and 10 drug-like compounds were selected, out of which procurement and pKa measurements for 17 fragment-like and 7 drug-like compounds were successful, respectively. (B) Frequent heterocycles found in FDA approved kinase inhibitors, as determined by Bemis-Murcko fragmentation into rings [49]. Black structures were represented in SAMPL6 set at least once. Compounds with piperazine and indazole (gray structures) could not be included in the challenge set due to library and selection limitations. (C) Structures of heterocycles that were overrepresented based on our compound selection workflow. We have limited the number of occurrences of these heterocycles to at most one.