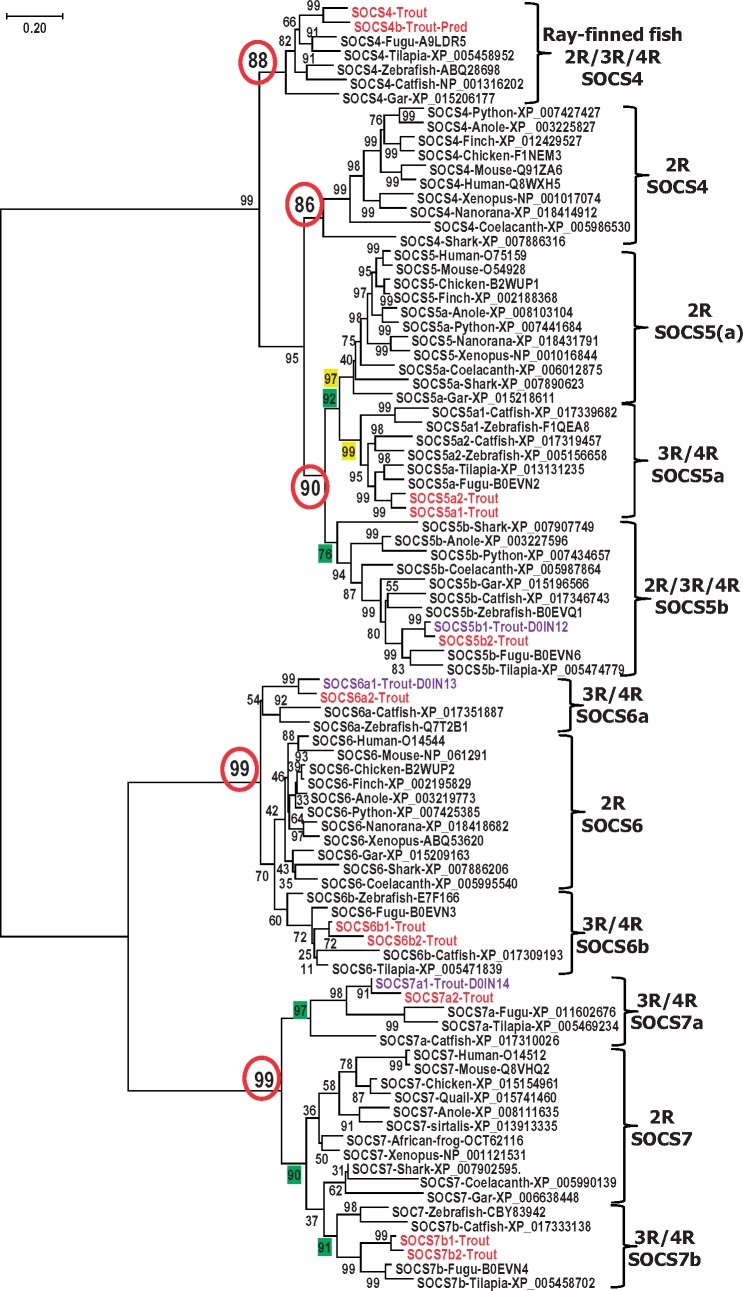
Fig. 3.
Phylogenetic tree analysis of vertebrate type I SOCS (SOCS4–7). The phylogenetic tree was constructed using amino acid multiple alignments generated by ClustalW and the Neighbor-Joining method within the MEGA7 program. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (10,000 replicates) is shown next to the branches. The evolutionary distances were computed using the JTT matrix-based method with the pairwise deletion option. The accession number of each sequence is shown after the common species name. The trout sequences reported in this study are in red and those known previously in purple.