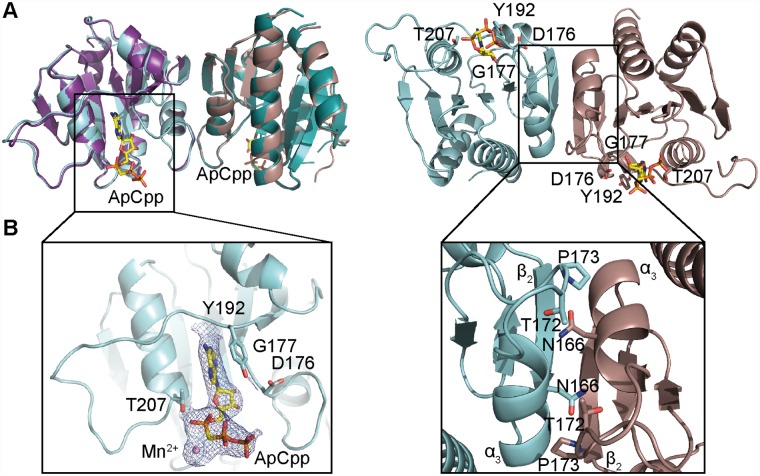
Fig 1. Crystal structure of the S. aureus DacACD protein in the nucleotide-free and nucleotide-bound states.
(A) Protein structure of the S. aureus DacACD protein with and without ApCpp, shown in ribbon representation. The apo S. aureus DacACD structure was solved by molecular replacement using the L. monocytogenes CdaA protein (PDB 4RV7) as template. The two monomers in the crystallographic unit are shown in teal and purple (for apo-DacACD) and in cyan and brown (for ApCpp-DacACD). The dimer is shown in two views with the second view in a 90° angle with the predicted active site residues D176, G177, T207 shown in stick representation in the 90° view (right). While the dacA sequence starting from codon F101 were included in the construct, only residues Y110 to G260 were visible in chain A and residues S111 to T261 in chain B. (B) Zoomed-in view of the S. aureus ApCpp-DacACD binding site (left) and DacACD dimerization interface (right). Electron densities corresponding to ApCpp and the metal ion are displayed. ApCpp and the DacACD interacting residues are shown in stick representation. Amino acid residues N166, T172 and P173 providing hydrogen bonding or van der Waals interactions are also shown in stick representation.