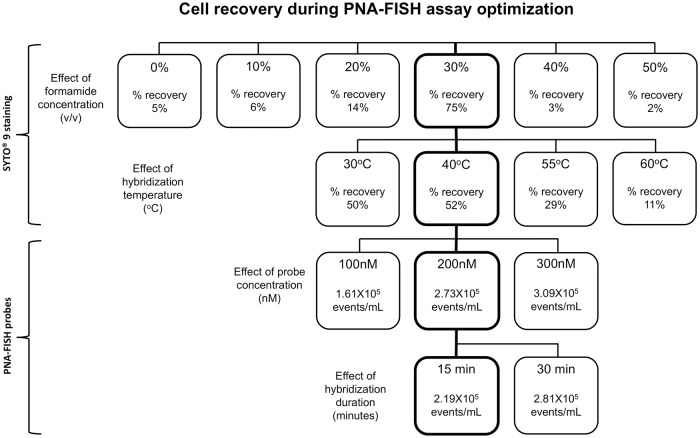
Fig 3. PNA-FISH assay optimisation flowchart using pure bacterial culture.
Four optimisation protocols were performed one of each for formamide concentration, hybridization temperature, probe concentration, and hybridization duration. In the first two protocols (top rows), SYTO 9 was used to determine changes in the overall bacterial population subjected to variations in formamide concentrations and temperature. Initially, varying concentrations of formamide were added to bacterial suspensions containing equal amounts of bacteria followed by incubation at 40°C for 15 minutes prior to staining with SYTO 9. The highest bacterial recovery of 75% was achieved with 30% formamide (v/v, thick frame) compared to other treated and untreated samples. This formamide concentration (30%) was selected for subsequent experiment in which bacterial cells were exposed to varied hybridization temperatures from 30°C to 60°C (second row). The best result with 52% bacterial cell recovery was obtained at 40°C (thick frame). For the last two optimisation steps, PNA-FISH probe was used in hybridization buffer containing 30% formamide at 40°C (third and fourth rows). In the third row, three different probe concentrations from 100 to 300 nM were tested by PNA-FISH. The probe concentration of 200 nM turned out to be optimal (thick frame) since no significant increase in hybridisation signal was observed when the probe concentration was elevated to 300 nM. Eventually, hybridization duration of 15 minutes was determined as optimal since the majority of the hybridization positive events were detected in the first 15 minutes. The squares with thick frames stacked vertically indicate optimised hybridization conditions applied to all subsequent experiments.