SUMMARY
Biological systems must possess mechanisms that prevent inappropriate responses to spurious environmental inputs. Caenorhabditis elegans has two breakdown pathways for the short-chain fatty acid propionate: a canonical, vitamin B12-dependent pathway and a propionate shunt that is used when vitamin B12 levels are low. The shunt pathway is kept off when there is sufficient flux through the canonical pathway, likely to avoid generating shunt-specific toxic intermediates. Here, we discovered a transcriptional regulatory circuit that activates shunt gene expression upon propionate buildup. Nuclear hormone receptor 10 (NHR-10) and NHR-68 function together as a “persistence detector” in a type 1, coherent feed-forward loop with an AND-logic gate to delay shunt activation upon propionate accumulation and to avoid spurious shunt activation in response to a non-sustained pulse of propionate. Together, our findings identify a persistence detector in an animal, which transcriptionally rewires propionate metabolism to maintain homeostasis.
Graphical Abstract
In Brief
Bulcha et al. discover a transcriptional persistence detector composed of a type 1 coherent feed-forward loop with an AND-logic gate that rewires propionate metabolism in C. elegans.
INTRODUCTION
Propionate is a three-carbon, short-chain fatty acid that is generated by the breakdown of odd-chain fatty acids and branched-chain amino acids. Propionate is toxic when it accumulates and is eliminated from the body by a breakdown pathway that uses vitamin B12 as a co-factor (Deodato et al., 2006).
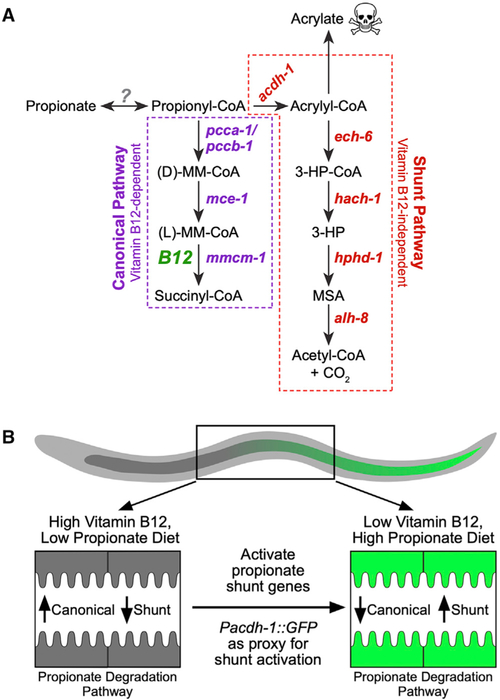
The nematode Caenorhabditis elegans is a bacterivore that can grow on a variety of diets that can be high or low in vitamin B12. This metabolic flexibility is enabled in part because C. elegans has two propionate breakdown pathways: the canonical vitamin B12-dependent pathway and an alternate pathway, or shunt, that is used under low vitamin B12 dietary conditions or when flux through the canonical pathway is genetically perturbed (Watson et al., 2013, 2014, 2016) (Figure 1A). The cooccurrence of two parallel pathways begs the question of why the animal has maintained the canonical propionate breakdown pathway. In other words, why not just use the propionate shunt? When C. elegans is grown on bacterial diets with high levels of vitamin B12, expression of the five propionate shunt genes is low, with the first gene in the pathway, acdh-1, being almost off (Watson et al., 2014, 2016). This indicates that the animal strongly prefers to use the canonical pathway and prevents activation of the shunt pathway when it is not needed.
Figure 1. Two C. elegans Propionate Breakdown Pathways That Function in the Animal’s Intestine.
(A) Cartoon of C. elegans propionate breakdown pathways. B12, vitamin B12; 3-HP, 3-hydroxypropionate; MM-CoA, methylmalonyl-CoA; MSA, malonic semialdehyde.
(B) Cartoon of the activation of propionate shunt expression in the 20-cell C. elegans intestine under different dietary conditions.
One reason for keeping flux through the propionate shunt pathway low when it is not needed is that intermediates in this pathway can be toxic when they accumulate. For instance, the first step in the propionate shunt involves the conversion of propionyl-coenzyme A (CoA) into acrylyl-CoA, which can be converted into the highly toxic intermediate acrylate upon dissociation of the CoA. Animals in which ech-6, encoding the enzyme that metabolizes acrylyl-CoA (Figure 1A), is perturbed are very sick, whereas a double perturbation of ech-6 and acdh-1, encoding the enzyme that generates acrylyl-CoA, rescues this sickness, as the perturbation of acdh-1 prevents the production of acrylate in the first place (Watson et al., 2016) (Figure 1A).
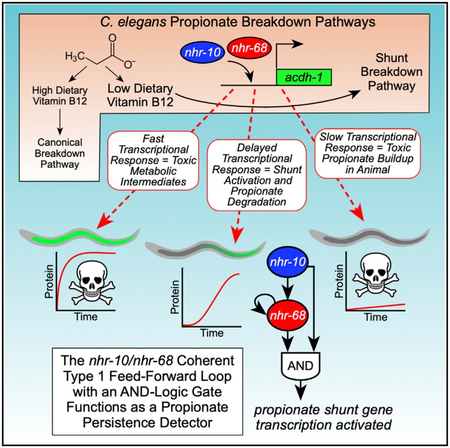
How does C. elegans ensure that the propionate shunt is kept off and is only activated when it is really needed? Here, we identify two nuclear hormone receptors (NHRs), nhr-10 and nhr-68, that are both transcriptionally and functionally important for the activation of shunt gene expression in response to the excessive accumulation of propionate. We find that nhr-10 activates nhr-68 and that nhr-68 expression alone is not sufficient to drive propionate shunt gene activation, i.e., both nhr-10 and nhr-68 are required, indicating that they do not act in a simple linear pathway. Together, our findings indicate that nhr-10 and nhr-68 function together in a gene regulatory network circuit known as a feed-forward loop (FFL) with an AND-logic gate. Previous modeling by Uri Alon has led to the specific prediction that FFLs with AND-logic gates will generate a delay in target gene activation and that a short pulse of input is not sufficient to activate target gene expression, and, therefore, such circuits have been named “persistence detectors” (Alon, 2007). However, with the exception of the L-arabinose utilization system in Escherichia coli (Mangan and Alon, 2003), the existence and importance of transcriptional persistence detector circuits in multicellular organisms has remained unclear. We demonstrate that the nhr-10/nhr-68 circuit functions as a persistence detector in several ways. First, there is a ~3-hr delay in propionate shunt activation upon the supplementation of propionate. Second, a 1-hr pulse of propionate is not sufficient to activate propionate shunt gene expression. Finally, we show that NHR-68 overexpression is not sufficient to activate propionate shunt gene expression in response to propionate, demonstrating that the two NHRs do not function in a simple linear pathway. We propose that the propionate persistence detector functions to ensure that the propionate shunt stays off unless propionate accumulation is persistent, thereby preventing the unwanted generation of highly toxic shunt intermediates. This gene regulatory network architecture links dietary input to metabolic output to ensure animal homeostasis.
RESULTS
The Nuclear Hormone Receptors nhr-10 and nhr-68 Activate Propionate Shunt Gene Expression in Response to Propionate
Propionate is generated and broken down in the C. elegans intestine, an organ composed of precisely 20 cells that functions both as the animal’s gut and its liver (Figure 1B). Our previous data indicate that the animal strongly prefers to use the canonical, vitamin B12-dependent propionate breakdown pathway and that it has tight control mechanisms to keep the propionate shunt pathway off unless it is needed, such as under persistently low vitamin B12 conditions. We have used a transgenic C. elegans strain that expresses the GFP under the control of the acdh-1 gene promoter as a proxy for shunt gene expression (Arda et al., 2010; MacNeil et al., 2013; Watson et al., 2013, 2014, 2016). acdh-1 encodes an acyl-CoA dehydrogenase that catalyzes the first reaction in the propionate shunt pathway and is most highly expressed in the C. elegans intestine (Arda et al., 2010; MacNeil et al., 2013; Watson et al., 2013, 2014, 2016). When these transgenic animals are fed a bacterial diet that is low in vitamin B12, intestinal GFP expression is high, whereas GFP levels are low on diets high in vitamin B12 (MacNeil et al., 2013; Watson et al., 2014, 2016) (Figure 1B). In addition, GFP expression is activated when genes in the canonical propionate breakdown pathway are genetically perturbed, even in the presence of vitamin B12, or when propionate is supplemented to high vitamin B12 bacterial diets (Watson et al., 2013, 2014, 2016). We have previously used the Pacdh-1::GFP strain in the context of defining a C. elegans intestinal gene regulatory network by comprehensive transcription factor (TF) RNAi, and found more than 40 TFs that activate the acdh-1 promoter in the absence of exogenously supplemented propionate (MacNeil et al., 2015). However, it is not clear whether all or only a subset of these TFs specifically mediate the transcriptional response to propionate.
To identify the TFs that specifically activate acdh-1 expression in response to propionate, we tested the previously found TFs in the presence of both vitamin B12 (to repress basal GFP expression) and propionate (to specifically activate GFP expression) (Figure S1A). Interestingly, we found that RNAi of only a subset of the 43 TFs that affected GFP expression under untreated conditions also affected GFP levels on propionate supplemented conditions (Figures 2A and S1B). Specifically, RNAi of 16 TFs reduced GFP expression under both conditions, RNAi of 26 TFs only repressed GFP expression on untreated conditions, and RNAi of one TF, mxl-3, reduced GFP expression on untreated conditions but activated the acdh-1 promoter on propionate-supplemented conditions. These results indicate that the acdh-1 promoter not only responds to excess propionate but to other cellular conditions as well and that the response to these other conditions involves other TFs.
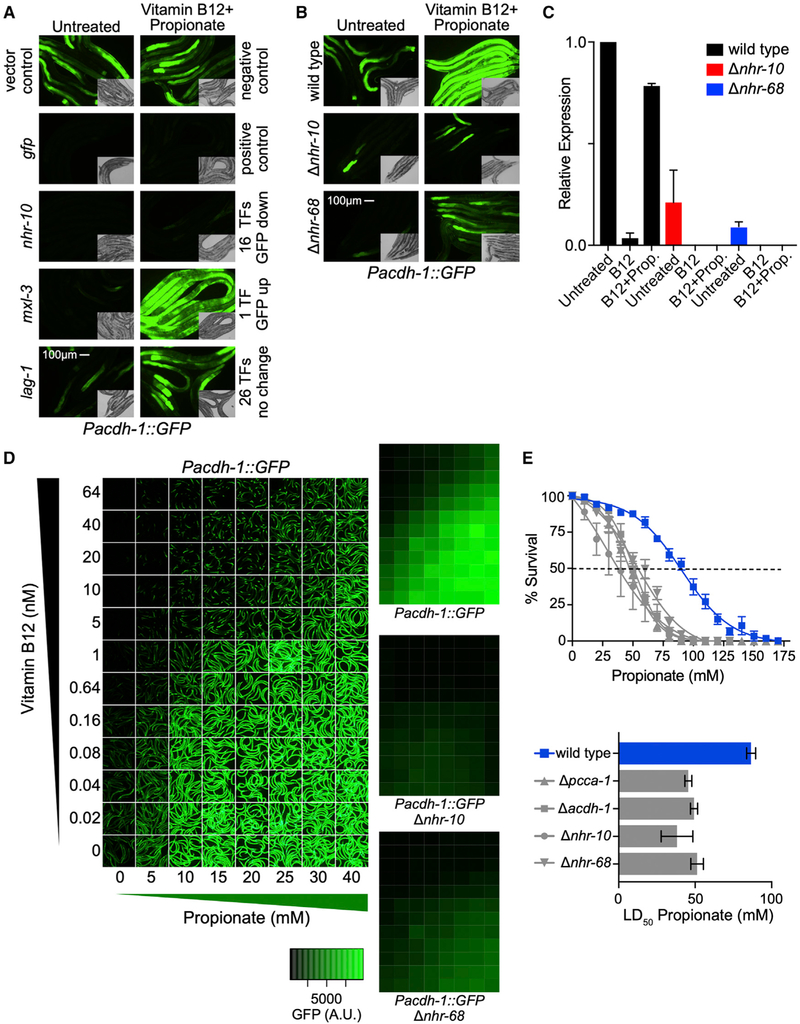
Figure 2. The Nuclear Hormone Receptors NHR-10 and NHR-68 Mediate the Transcriptional and Functional Response to Excess Propionate.
(A) Fluorescent microscopy images of TF RNAi show that only a subset of TFs that activate the acdh-1 promoter in animals fed E. coli HT115 bacteria are involved in the transcriptional response to excess propionate. Insets show differential interference contrast (DIC) images.
(B) The transcriptional response to excess propionate is greatly reduced in nhr-10 and nhr-68 deletion mutants, validating the TF RNAi results.
(C) qRT-PCR experiment showing that endogenous acdh-1 expression is not induced in response to propionate in nhr-10 and nhr-68 deletion mutants.
(D) nhr-10 and nhr-68 are required for acdh-1 promoter activation in a broad range of vitamin B12 and propionate concentrations. Left panel shows images of Pacdh-1::GFP animals supplemented with indicated concentrations of vitamin B12 and/or propionate. Right three panels show quantification of GFP levels at each concentration in Pacdh-1::GFP animals in wild-type, nhr-10, or nhr-68 deletion mutant animals. Quantification is the average of nine experiments: three biological replicates with three technical replicates each.
(E) Propionate toxicity assays show that nhr-10 and nhr-68 are functionally required to mitigate the toxic effects of propionate. Top panel shows propionate dose-response curves, and bottom panel shows LD50 values.
Several of the 16 TFs that reduce GFP expression when knocked down by RNAi function at high levels in the intestinal gene regulatory network and likely affect acdh-1 promoter activity indirectly (MacNeil et al., 2015). For instance, the intestinal master regulator elt-2 broadly controls intestinal gene expression and resides at the top of the hierarchy (MacNeil et al., 2015; McGhee et al., 2007). Knock down of either of the two TFs, nhr-10 or nhr-68, had a strong effect on acdh-1 promoter activity under propionate-supplemented conditions, and these TFs reside low in the gene regulatory network hierarchy (MacNeil et al., 2015). As TFs that reside low in the hierarchy tend to directly affect the promoter, this result suggests that nhr-10 and nhr-68 may be critical for propionate shunt activation. Indeed, deletion of either nhr-10 or nhr-68 greatly reduced acdh-1 promoter activity, as well as endogenous acdh-1 expression (Figures 2B and 2C). Quantitative analysis of GFP expression in a broad range of vitamin B12 and propionate concentrations revealed that nhr-10 and nhr-68 are both required for acdh-1 promoter activation under a broad range of propionate concentrations. Although nhr-10 is absolutely required, there is still modest GFP expression in nhr-68 deletion mutant animals (Figure 2D). These observations indicate that nhr-10 and nhr-68 together activate the expression of propionate shunt gene expression in response to excess propionate.
Are nhr-10 and nhr-68 functionally important to prevent the buildup of toxic propionate? We previously found that acdh-1 expression under low vitamin B12 conditions is important to mitigate the effects of excess propionate (Watson et al., 2014, 2016). Specifically, the lethal dose 50 (LD50) of propionate in acdh-1 mutant animals fed an E. coli diet low in vitamin B12 is similar to that of pcca-1 deletion mutants, in which flux through the canonical propionate breakdown pathway is perturbed (Figures 1A and 2E) (Watson et al., 2014, 2016). Here, we found that both nhr-10 and nhr-68 deletion mutants are more sensitive to excess propionate than wild-type animals (Figure 2E). Importantly, deletion of either gene renders the animals equally sensitive to propionate as deletion of their transcriptional target acdh-1, which shows that both of these TFs are functionally important to mitigate propionate toxicity. Altogether, these data show that nhr-10 and nhr-68 are both required for transcriptional as well as functional activation of propionate shunt gene expression.
nhr-10 and nhr-68 Activate All Five Propionate Shunt Genes in Response to Excess Propionate
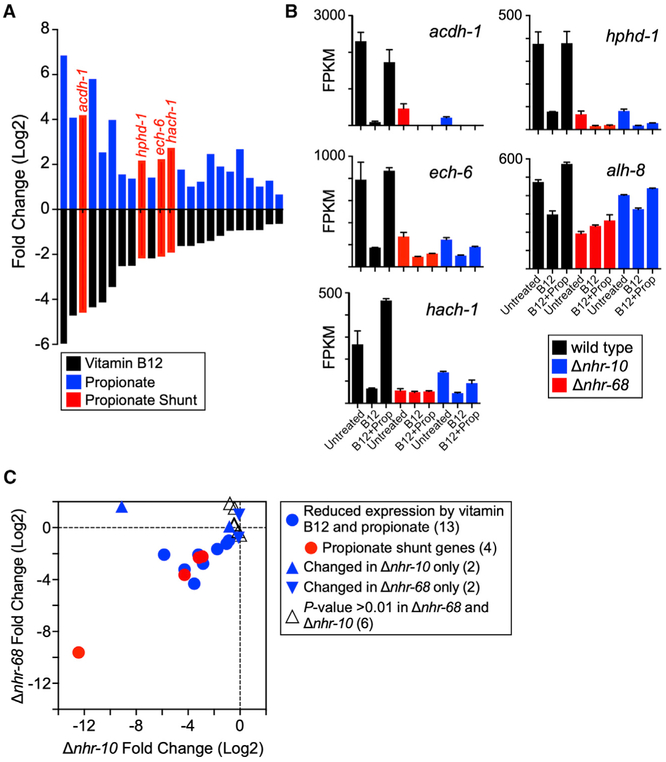
So far, we have used the Pacdh-1::GFP transgenic strain as a proxy for studying propionate shunt activation. To investigate activation of the other four propionate shunt genes and to identify additional genes that are repressed by vitamin B12 and activated by propionate, we performed RNA sequencing (RNA-seq) on wild-type, Δnhr-10, and Δnhr-68 mutant animals under untreated, vitamin B12 only, and vitamin B12 + propionate-supplemented conditions. We identified 23 genes that in wild-type animals are repressed by vitamin B12 and activated by propionate, including four of the five propionate shunt genes (Figure 3A). The fifth gene alh-8 behaved similarly with an adjusted p value <0.01 but was just below our cutoff of a fold-change greater than or equal to 1.5 (Figure 3B; Table S1). Importantly, activation of 13 of these 23 genes required both nhr-10 and nhr-68, including the propionate shunt genes (Figure 3C). This indicates that a larger gene battery than just the propionate shunt genes is under control of both NHRs and indicates that additional genes may be involved in propionate shunt metabolism or regulation.
Figure 3. Expression Profiling of nhr-10 and nhr-68 Deletion Mutant Animals.
(A) Bar graph showing the 23 genes that are significantly repressed by vitamin B12 and induced by propionate in wild-type animals, as identified by RNA-seq.
(B) Bar graph of RNA-seq fragments per kilobase of transcript per million mapped reads (FPKM) data showing that all five propionate shunt genes are activated by nhr-10 and nhr-68 in response to excess propionate.
(C) Scatterplot showing that 13 of the 23 genes repressed by vitamin B12 and induced by propionate are controlled by both nhr-10 and nhr-68. Gene numbers for each condition are in parentheses.
nhr-10 and nhr-68 Function in a Coherent Type 1 FFL with an AND-Logic Gate
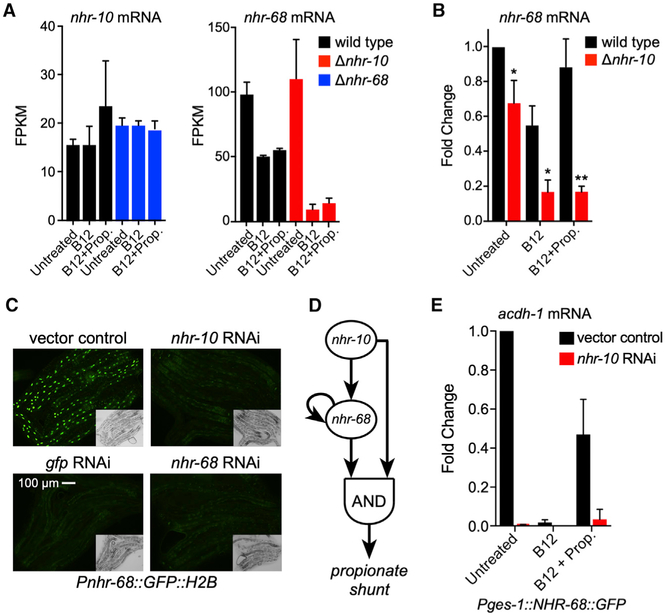
Although nhr-10 mRNA levels are not affected by vitamin B12 or propionate supplementation, nhr-68 expression is repressed by vitamin B12 and activated by propionate (Figures 4A and 4B). Interestingly, we found that nhr-68 mRNA levels are reduced in nhr-10 deletion mutant animals both by RNA-seq and by qRT-PCR (Figures 4A and 4B), which indicates that nhr-10 activates the expression of nhr-68. RNAi of nhr-10 in Pnhr-68::GFP::H2B transgenic animals led to a reduction in GFP expression compared to vector control RNAi, validating this result (Figure 4C). Interestingly, nhr-68 RNAi also caused a reduction in GFP levels in this strain, indicating that nhr-68 activates its own expression. Altogether, our observations indicate that nhr-10 and nhr-68 function in a specific type of circuit known as a coherent type 1 FFL with an AND-logic gate (Figure 4D). However, our data so far do not exclude the possibility that nhr-10 and nhr-68 function in a simple linear pathway where nhr-10 activates nhr-68 and nhr-68 activates propionate shunt gene expression. To test this idea, we overexpressed nhr-68 under the control of the promoter of moderately and highly constitutively expressed intestinal genes ges-1 and asp-5, respectively. Neither of these genes is affected by vitamin B12 or propionate or by deletion of nhr-10 (Figures S2A and S2B). We examined the induction of acdh-1 expression by qRT-PCR either with vector control or with nhr-10 RNAi in both strains. We found that, in these animals, acdh-1 mRNA levels were repressed by vitamin B12 and induced by propionate, just as in wild-type animals (Figures 4E and S2C). However, acdh-1 levels were greatly reduced upon RNAi of nhr-10, indicating that nhr-10 is absolutely required for propionate shunt activation. This finding demonstrates that activation of nhr-68 by nhr-10 is not sufficient for propionate shunt gene induction in response to propionate but rather that the path leading from nhr-10 to these genes is essential as well. This essential role of nhr-10 is in agreement with our previous observation that this TF directly binds the acdh-1 gene promoter (Arda et al., 2010; MacNeil et al., 2015). These observations indicate that nhr-10 and nhr-68 do not function in a simple linear genetic pathway but that they function in a more complex circuitry, represented by the type 1 coherent FFL in Figure 4D.
Figure 4. nhr-10 Activates nhr-68 Expression and nhr-68 Is an Autoactivator.
(A) Bar graph of RNA-seq data for nhr-10 and nhr-68 mRNA levels in the absence of nhr-68 (left) and nhr-10 (right).
(B) Bar graph showing qRT-PCR data for nhr-68 mRNA levels in the absence of nhr-10. Statistical differences between wild-type and nhr-10 deletion mutants were determined by two-tailed paired Student’s t test (*p < 0.05, **p < 0.005).
(C) Fluorescent microscopy images of RNAi of nhr-10 or nhr-68 shows reduced GFP expression in Pnhr-68::GFP::H2B transgenic animals. Insets show DIC images.
(D) Cartoon of the nhr-10/nhr-68 gene regulatory network circuit.
(E) qRT-PCR shows that constitutive intestinal expression of NHR-68 under the control of the ges-1 promoter does not induce acdh-1 expression in response to propionate. All measurements are statistically significantly different compared to untreated vector control as determined by two-tailed paired Student’s t test (p < 0.05).
The nhr-10/nhr-68 Coherent Type 1 FFL with an AND-Logic Gate Functions as a Propionate Persistence Detector
Mathematical modeling of type 1 coherent FFLs with AND-logic gates has led to the prediction that such circuits function as persistence detectors that generate a delay in target gene expression and only activate downstream target genes when the input is sustained. This is because it takes time for the first TF in the circuit to activate the second and both are required (Alon, 2007). However, this prediction has so far only been experimentally substantiated using a synthetic circuit in E. coli (Mangan and Alon, 2003).
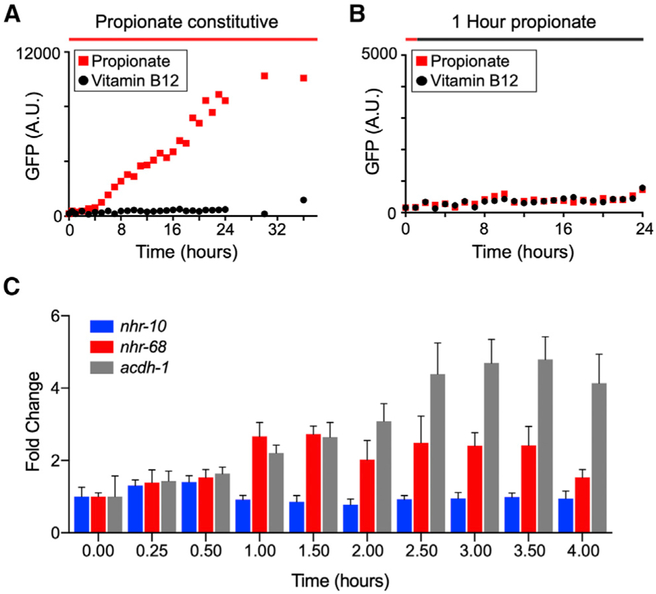
Next, we asked whether the nhr-10/nhr-68 circuit may function as a genuine persistence detector. To do so, we tested two predictions based on previous modeling (Mangan and Alon, 2003): first that activation of acdh-1 would exhibit a delay upon propionate supplementation, and second that a short pulse of propionate supplementation would not induce acdh-1 activation. We first constitutively supplemented Pacdh-1::GFP transgenic animals kept on vitamin B12 (GFP expression off) with propionate and quantified GFP expression every 30 min the first 2 hr and every hr for 22 hr and after 30 and 36 hr. As expected, GFP stayed off in animals that were constitutively supplemented with vitamin B12 (Figure 5A). When supplemented with propionate, however, GFP expression was robustly induced. Importantly, this induction occurred with a delay of ~3 hr. When Pacdh-1::GFP animals were given the same concentration of propionate, but only in a 1-hr pulse, GFP expression was not induced (Figure 5B). Finally, using qRT-PCR of endogenous nhr-10, nhr-68 and acdh-1 expression, we again found that nhr-10 expression does not change with propionate, whereas nhr-68 expression is modestly induced after about 1 hr, after which it stabilizes, followed by a longer induction of acdh-1 mRNA. Note that the induction of the acdh-1 mRNA is a bit faster than the induction of GFP expression in the reporter strain and that it tapers off, likely because of mRNA decay.
Figure 5. acdh-1 Expression Is Induced with a 3-Hr Delay in Response to Propionate and Does Not Respond to a 1-Hr Propionate Pulse.
(A) GFP expression in Pacdh-1::GFP animals transferred from vitamin B12 (black circles) to constitutive propionate supplementation (red squares) shows a ~3-hr delay.
(B) A 1-hr pulse of propionate does not induce GFP expression in Pacdh-1::GFP animals.
(C) qRT-PCR experiment of endogenous nhr-10, nhr-68 and acdh-1 expression upon propionate supplementation.
nhr-68 Autoactivation Can Modulate the Delay in Persistence Detector Target Gene Activation
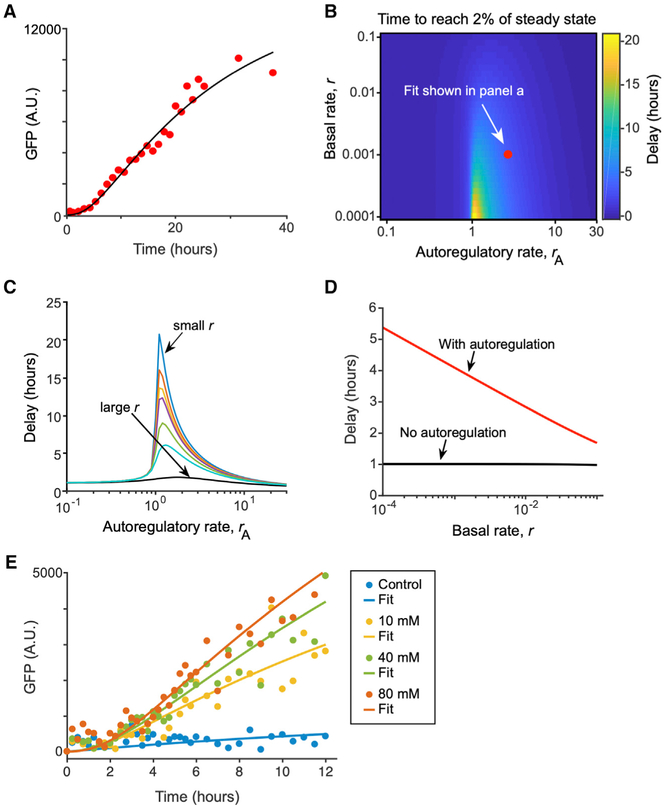
Next, we asked how nhr-68 autoactivation may contribute to the functionality of the persistence detector. For this, we modeled a type 1 coherent FFL with an AND-gate with autoregulation of nhr-68 and simulated the system using Michaelis-Menten kinetics. We used the fitted experimental data from Figure 5A to estimate model parameters (Figure 6A; see STAR Methods). We then explored the interplay between nhr-68 basal expression rate (characterized by the parameter, r) and nhr-68 autoregulatory expression rate (characterized by rA) on the observed delay in target gene expression. Figure 6B shows a heatmap of delay times as a function of these two parameters over a broad range of values. Although precise quantitative values of the model parameters are not known, we found an interesting general feature of the circuit. Without autoactivation, the system has a tight range of predicted delay times that is relatively insensitive to the basal rate of induction and is set by the decay rates of the constituent TFs (Figure 6C). However, the inclusion of autoregulation of nhr-68 enables a wide range of delays that are set by the basal expression rate (which, in our model, is controlled solely by nhr-10). In other words, small basal rates resulting from lower input signals would trigger longer delays in target gene expression, whereas strong input signals would trigger shorter delays and a quicker response (Figures 6C and 6D). To test this prediction, we supplemented Pacdh-1::GFP animals with different concentrations of propionate and examined the induction of GFP expression. Indeed, we found that higher concentrations of sustained propionate result in faster activation of GFP expression (Figure 6E).
Figure 6. Computational Modeling Shows That nhr-68 Autoactivation Can Modulate the Delay in Target Gene Expression.
(A) Fit of propionate supplementation data from Figure 5A to estimate approximate parameter range.
(B) Heatmap of delay time to reach 2% of steady state level of GFP based on the rate of basal expression, r, and the rate of autoregulation, rA, of the nhr-68 gene.
(C) Delay time as a function of autoregulatory strength for several basal rates from the range of values examined in (B).
(D) Delay time with autoregulation (using rA found from C) and without autoregulation as basal rate is tuned.
(E) Dynamics of Pacdh-1::GFP activation in response to different propionate concentrations.
Taken together, propionate shunt activation only occurs when propionate buildup is sustained and requires a transcriptional circuit involving nhr-10 and nhr-68 that function as a persistence detector in a type 1 coherent FFL with an AND-logic gate.
DISCUSSION
We have discovered a transcriptional persistence detector in a multicellular organism. This persistence detector activates the five genes comprising the propionate shunt pathway in C. elegans. This pathway provides an alternative way to catabolize propionate, which is toxic when it accumulates in both humans and C. elegans (Deodato et al., 2006; Watson et al., 2016). Shunting of propionate occurs in both humans and C. elegans, as indicated by the detection of shunt pathway intermediates in propionic acidemia patients and individuals with mutations in other relevant genes (Ando et al., 1972; Deodato et al., 2006; Peters et al., 2015). However, although C. elegans has evolved a propionate shunt pathway that is transcriptionally induced when propionate accumulates, humans likely repurpose metabolic enzymes that function in other metabolic pathways (Watson et al., 2016).
Two observations indicate that C. elegans preferentially uses the canonical, vitamin B12-dependent propionate breakdown pathway, rather than the propionate shunt. First, the canonical pathway has not been lost in evolution, which would be expected if its function became obsolete. Second, the shunt pathway is inactive when flux through the canonical pathway is enabled by the sufficient dietary intake of vitamin B12. One reason for the preferential use of the canonical pathway is that the propionate shunt generates acrylate, which is much more toxic than propionate. Acrylate is produced in the first reaction in the propionate shunt where propionyl-CoA is converted into acrylyl-CoA (which can be interconverted with acrylate when the CoA is chemically or enzymatically removed) by the ACDH-1 enzyme (Figure 1). The next enzyme in the shunt pathway, ECH-6, then converts acrylyl-CoA into 3-hydroxypropionyl-CoA. We previously found that RNAi of ech-6 renders the animals very sick and that double perturbation of ech-6 with acdh-1 suppresses this phenotype (Watson et al., 2016). Our data indicate that C. elegans ensures that the shunt pathway stays off until it is really needed by using a transcriptional persistence detection mechanism.
The activation of shunt gene expression, as measured using the Pacdh-1::GFP reporter strain, occurs with a delay of ~3 hr. This suggests that the response to excess propionate is on a relatively long timescale due to a relatively straightforward metabolite buildup rather than as a short-acting signal. Although our study illuminates the system-level mechanism of propionate persistence detection, the precise molecular mechanism remains to be elucidated. We previously found that NHR-10 physically binds the acdh-1 promoter in yeast one-hybrid assays (Arda et al., 2010; MacNeil et al., 2015). We do not yet know whether it also interacts with the other shunt gene promoters. One possibility is that NHR-68 and NHR-10 physically interact to form a heterodimer. However, we did not detect any interactions with NHR-68 in our large-scale protein-DNA and protein-protein interaction studies (Fuxman Bass et al., 2016; Reece-Hoyes et al., 2013), so this remains to be investigated. We also do not yet know the mechanism of propionate detection by the persistence detector and how much intracellular propionate buildup is required to activate the circuit. Propionate is generated from odd-chain fatty acids, branched-chain amino acids, methionine, and threonine (Yilmaz and Walhout, 2016), and is catabolized in the mitochondria (Al-Lahham et al., 2010). How information involved in propionate shunt activation is transferred from the mitochondria and cytoplasm to NHRs in the nucleus is not known. It is tempting to speculate that NHR-10, NHR-68, or both directly interact with propionate, a three-carbon short-chain fatty acid, given that NHRs are known to use fatty acids as ligands (Evans and Mangelsdorf, 2014). However, the small size and volatility of propionate make detection of its interaction with proteins extremely challenging.
Several observations indicate that the persistence detector does not function in isolation and does not function solely to activate the expression of acdh-1 and other genes but, rather, that it is embedded in a larger gene regulatory network. First, the set of downstream targets consists of at least 13 genes that are repressed by vitamin B12, activated by propionate, and dependent on both nhr-10 and nhr-68. Aside from four shunt genes, this set contains nine genes, the function of which is largely unknown. It is likely that several of these genes function to support shunt function or to enable its shutdown when nutritional conditions favor the use of the canonical vitamin B12-dependent propionate breakdown pathway. Second, many additional TFs are involved in acdh-1 expression (MacNeil et al., 2015; this study). These include 16 TFs that, when knocked down by RNAi, reduce acdh-1 promoter activity either induced by propionate supplementation or under untreated conditions. Most of these TFs have a partial effect and reside higher in the hierarchy of the intestinal gene regulatory network (MacNeil et al., 2015) and likely affect the acdh-1 promoter indirectly. RNAi of one TF, mxl-3, reduced acdh-1 promoter activity on E. coli bacteria but increased it under propionate-supplemented conditions. We did not follow up on this observation because we observed only very small effects on propionate shunt gene expression in mxl-3 mutant animals (data not shown). Another set of 26 TFs regulate acdh-1 under untreated conditions only, i.e., their knockdown has no effect on propionate-supplemented conditions. This finding indicates that acdh-1 responds to other metabolites that act by other TFs. Interestingly, these TFs include nhr-101 and nhr-114, which would be appealing candidates to mediate the response to other metabolites. It is interesting to note that acdh-1 expression is not completely off in either nhr-10 or nhr-68 deletion mutant in the untreated condition (Figure 2D) and that this residual expression is repressed by vitamin B12. This observation suggests that other metabolites activating acdh-1 may also be functionally connected to vitamin B12 metabolism. Moreover, it suggests that acdh-1 may have a function outside the propionate shunt. Taken together, we discovered a persistence detector in a multicellular organism and linked this gene regulatory network architecture to a functional metabolic response in a whole animal.
STAR★METHODS
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for reagents may be directed to and will be fulfilled by the Lead Contact, A.J.M. Walhout (marian.walhout@umassmed.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
C. elegans Strains
N2 (Bristol) was used as the wild-type strain, and animals were maintained on nematode growth medium (NGM) at 20°C as described (Brenner, 1974). Transgenic strains VL1286 (wwSi28[Pnhr-68::GFP::H2B; unc-119(+) II]), VL1296 (wwSi29[Pges-1::NHR-68::GFP; unc-119(+) II]), VL1297 (wwSi30[Pasp-5::NHR-68; unc-119(+) II]) were developed by using the Mos1-mediated single-copy insertion (MosSCI) method (Frøkjær-Jensen et al., 2014) and the integrated transgenes were confirmed by PCR genotyping (Table S2).
METHOD DETAILS
RNA Interference
RNAi was performed as described (MacNeil et al., 2015) with or without supplementation of 5 nM vitamin B12 and 40 mM propionate. Changes in intestinal GFP were scored visually when samples contained a mix of L4 and young adult animals. Knockdowns were scored as positive when most animals in the well displayed a change in intestinal GFP. Changes in GFP levels in other tissues were not recorded. Experiments were performed five independent times. TFs that scored in at least three independent experiments were considered hits. All RNAi clones included in the final dataset were sequence-verified.
Propionate and Vitamin B12 Gradient Assays
L1 synchronized animals were grown on NGM media supplemented with a matrix of propionate (0 to 40 mM) and vitamin B12 (0 to 64 nM) concentrations. GFP fluorescence was measured using a Tecan Infinite M1000Pro microplate reader as described (Leung et al., 2011). Five adult animals were randomly picked in triplicate and transferred to a 384 well plate containing 35 μl of buffer (M9 buffer, 1 μM levamisole and 0.5% PEG). GFP intensity measurement was performed at 485nm/9nm excitation and 535nm/20nm emission spectra. Each experiment was performed in biological triplicate with three technical triplicates each, and the fluorescence intensity of each biological replicate was averaged. The average fluorescence intensity was used to make the heatmap using the online tool http://www.heatmapper.ca/expression/.
Propionate Toxicity Assays
Propionate toxicity assays were performed as described previously (Watson et al., 2016). Approximately 100-200 synchronized L1 animals (hatched overnight, 20 hours post-bleach) were added to E. coli OP50-seeded 35 mm NGM agar plates containing various concentrations of pH-neutralized propionic acid. Each dose tested included two technical and three biological replicates. After 72 hours, animals that had developed past L1 stage were counted. The fraction surviving, S, as a function of propionic acid concentration, [C], was fit to the following dose response curve:
where S0 and S∞ are the fraction of animals surviving at zero and at saturating concentrations of propionic acid and n is the Hill slope. The dose required to kill 50% of the population, LD50, was found by least-squares fit of these four parameters. Toxicity assays were performed in biological triplicate, and the average LD50 was plotted ± SEM.
Expression Profiling
Animals were fed E. coli OP50 on NGM-agar supplemented with 10 nM vitamin B12. Animals were then grown on NGM-agar without vitamin B12 and synchronized for two generations by L1 arrest in M9 buffer for 18 hours post-treatment with buffered bleach. Animals were then grown on NGM-agar alone, or on NGM-agar supplemented with either 20 nM vitamin B12 or 20 nM vitamin B12 and 40 mM propionate. Approximately 3000 L4 stage animals were harvested and washed three times in M9 buffer for each condition. Total RNA was isolated using Trizol (Invitrogen) followed by DNase I (NEB) treatment and cleanup using Direct-zol RNA Mini Prep kit (Zymo Research). Two biological replicates for each condition were sequenced. Sequencing was performed by BGI using BGISEQ-500 platform with a single-end 50 bp read length, and a minimum of 26 million reads per sample. Differential gene expression between samples was analyzed using the standard output of BGI bioinformatics pipeline using EBSeq (Leng et al., 2013) for identifying differentially expressed genes. Differentially expressed genes were selected based on a fold change of ≥ ± 1.5, and adjusted P-value ≤ 0.01.
qRT-PCR
qRT-PCR was performed as described previously (Watson et al., 2016). Briefly, synchronized L1 animals were grown on NGM-agar plates containing 20 nM vitamin B12 and/or 40 mM propionate seeded with E. coli OP50 and grown at 20°C until they reached to late L4 stage. About 1500 animals were harvested for each condition, in triplicate. Animals were washed in M9 buffer and total RNA was isolated using TRIzol Reagent (Life Technologies), following by DNaseI (NEB) treatment and cleanup with Direct-zol RNA Mini Prep Kit (Zymo Research). cDNA was prepared from RNA using Oligo(dT) 12-18 Primer (Invitrogen) and M-MuLV Reverse Transcriptase (NEB). qPCR was performed in technical triplicate per gene condition using the Applied Biosystems StepOnePlus Real-Time PCR system and Fast Sybr Green Master Mix (ThermoFisher Scientific). Relative transcript abundance was determined by using the ΔΔCt method (18546601) and normalized to averaged ama-1 and act-1 mRNA expression levels. Primer sequences are provided in Table S2.
Propionate Pulse Experiments
L1 synchronized animals were grown on 10 nM vitamin B12 supplemented media seeded with E. coli OP50 until the adult stage. Animals were then transferred to 40 mM propionate supplemented media either constitutively, or for one hour, and transferred back to vitamin B12 supplemented media. During each transfer the animals were washed three times using M9 buffer. For each time point, five adult animals were randomly picked and transferred to a 384 well plate containing 35 μl M9 buffer containing 1 μM levamisole and 0.5% polyethylene glycol. GFP was measured at 485nm/20nm excitation and 535nm/20nm emission spectra as described above. Each experiment was performed in biological triplicate with three technical replicates each, and the fluorescence intensity of technical replicates were averaged for each biological replicate.
Modeling FFL with Positive Autoregulation
In the coherent FFL motif with an AND-logic gate (Figure 4D), propionate causes nhr-10 to activate nhr-68 expression with a basal rate r, and both nhr-10 and nhr-68 jointly activate GFP expression through an AND logic-gate with a rate rc. In the absence of the signal, rc = 0 due to the AND logic. Additionally, nhr-68 autoactivates with rate rA. Implicit in this model is the assumption that nhr-10 acts as a switch; i.e., nhr-10 is “on” in the presence of propionate and “off” in its absence, and that levels of nhr-10 do not change with time. To model the dynamics of the system we used Michaelis-Menten kinetics to arrive at the following ODEs that describe the system. GFP expression driven by the acdh-1 promoter in Pacdh-1::GFP transgenic animals was used as a proxy for propionate shunt expression:
where α is the degradation rate of nhr-68 or GFP and k is the dissociation constant. We assumed the degradation rate of nhr-68 to be relatively fast and set αnhr68 = 1 /hour. The decay rate of GFP, which is known to be very stable, was set to αGFP = 0.05/hour which is determined from fitting the decay of GFP signal over time when propionate is washed out; although αGFP does not have biological relevance, it impacts the dynamics of our measured signal and the rate of approach to steady state. The dissociation constant is set to 1. The steady state concentration of GFP is obtained by setting the ODEs to zero and solving for GFP and nhr-68 levels. The delay time, defined as the time to reach 2% of the maximum GFP level, is numerically obtained by solving the ODEs using the ODE solver, ODE45, in MATLAB. We fit the data for r, rA and rc in order to estimate the magnitude of these parameters. The role of autoregulation in the circuit was tested by varying the two contributions to nhr-68 production (autoregulation, characterized by rA and basal expression from nhr-10 alone, characterized by r) around these best-fit values. Adjusting the parameters k and αnhr68 alter the fit parameters, however, the highlighted qualitative features of the circuit remain unchanged.
QUANTIFICATION AND STATISTICAL ANALYSES
Error bars for FPKM numbers from RNA-seq experiments represent the standard deviation from the average of two experiments. Differentially expressed genes from the RNA-seq dataset were selected based on a fold change of 1.5 or more and a P-adjusted value of less than 0.01. The specific statistical parameters are represented in the figure legends of each figure.
DATA AND SOFTWARE AVAILABILITY
The RNA-sequencing data files were deposited in the NCBI Gene Expression Omnibus (GEO) under accession number GEO: GSE123507.
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Bacterial and Virus Strains | ||
| Escherichia coli OP150 | Caenorhabditis Genetics Center (CGC) | N/A |
| Escherichia coli HT115 | CGC | N/A |
| Escherichia coli HT115 Ahringer RNAi Library | (Kamath et al., 2003) | N/A |
| Escherichia coli HT115 ORFeome RNAi Library | (Rual et al., 2004) | N/A |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Propionic Acid | Sigma Aldrich | Cat#: P1386 |
| Adenosyl Cobalamin | Sigma Aldrich | Cat#: C0884 |
| BP clonase II | ThermoFisher Scientific | Cat#: 11789020 |
| Isopropyl β-D-1 thiogalactopyranoside (IPTG) | US Biological | Cat#: I8500 |
| Levamisole Hydrochloride | Sigma Aldrich | Cat#: PHR1798 |
| Polyethylene Glycol | Sigma Aldrich | Cat#: 202444 |
| TRIzol Reagent | Life Technologies | Cat#: 15596-018 |
| M-MuLV Reverse Transcriptase | NEB | Cat#: M0253 |
| Direct-zol RNA Mini Prep Kit | Zymo Research | Cat#: R2050 |
| DNase I | NEB | Cat#: M0303 |
| Oligo(dT) 12-18 Primer | Invitrogen | Cat#: 18418012 |
| RNaseOut | Invitrogen | Cat#: 10777019 |
| Fast SYBR Green Master Mix | ThermoFisher Scientific | Cat#: 4385616 |
| Deposited Data | ||
| Raw and analyzed RNA-seq data | This study | GEO: GSE123507 |
| Experimental Models: Organisms/Strains | ||
| Caenorhabditis elegans N2 (wild type) | CGC | N/A |
| C. elegans nhr-10(tm4695) | National Bioresource Project, Japan | WormBase: WBVar00253059 |
| C. elegans nhr-68(gk708) | CGC | Strain: VC1527 WormBase: WBVar00146032 |
| C. elegans pcca-1(ok2282) | CGC | Strain: RB1774 WormBase: WBVar00093442 |
| C. elegans acdh-1(ok1489) | CGC | Strain: VC1011 WormBase: WBVar00092700 |
| C. elegans wwIs24[Pacdh-1::GFP; unc-119(+)] | (MacNeil et al., 2013) | Strain: VL749 WormBase: WBTransgene00018139 |
| C. elegans nhr-10(tm4695); wwIs24[Pacdh-1::GFP; unc-119(+)] | (MacNeil et al., 2013) | Strain: VL868 |
| C. elegans nhr-68(tm708); wwIs24[Pacdh-1::GFP; unc-119(+)] | (Watson et al., 2013) | Strain: VL1113 |
| C. elegans wwSi28[Pnhr-68::GFP::H2B; unc-119(+) II] | This study | Strain: VL1286 |
| C. elegans wwSi29[Pges-1::NHR-68::GFP; unc-119(+) II] | This study | Strain: VL1296 |
| C. elegans wwSi30[Pasp-5::NHR-68; unc-119(+) II] | This study | Strain: VL1297 |
| Oligonucleotides | ||
| List of Oligonucleotides | This study | Table S2 |
| Recombinant DNA | ||
| pDONR P4-P1R | (Dupuy et al., 2007) | N/A |
| pDONR 221 | ThermoFisher Scientific | Cat#: 12536017 |
| pDONR P2R-P3 (for unc-54 3′ UTR) | N/A | |
| Software and Algorithms | ||
| StepOnePlus qPCR Software v2.3 | ThermoFisher Scientific | Cat#: 4376600 |
| MATLAB | Mathworks | https://www.mathworks.com/products/matlab.html |
| HeatMapper | (Babicki et al., 2016) | http://www.heatmapper.ca |
Highlights.
A transcriptional persistence detector activates propionate shunt only when needed
Persistence detection prevents generation of toxic shunt intermediates
nhr-10 and nhr-68 are persistence detectors in a feedforward loop with AND-logic gate
nhr-68 autoactivates and modeling shows that this can provide circuit tunability
ACKNOWLEDGMENTS
We thank members of the Walhout lab and the faculty in the Program in Systems Biology for discussion and critical reading of the manuscript. This work was supported by NIH grant DK068429 to A.J.M.W. Some bacterial and nematode strains used in this work were provided by the Caenorhabditis Genetics Center (CGC), which is funded by the NIH Office of Research Infrastructure Programs (P40 OD010440).
Footnotes
DECLARATION OF INTERESTS
The authors declare no competing interests.
SUPPLEMENTAL INFORMATION
Supplemental Information includes two figures and two tables and can be found with this article online at https://doi.org/10.1016/j.celrep.2018.12.064.
REFERENCES
- Al-Lahham SH, Peppelenbosch MP, Roelofsen H, Vonk RJ, and Venema K(2010). Biological effects of propionic acid in humans; metabolism, potential applications and underlying mechanisms. Biochim. Biophys. Acta 1801, 1175–1183. [DOI] [PubMed] [Google Scholar]
- Alon U (2007). Network motifs: theory and experimental approaches. Nat. Rev. Genet. 8, 450–61. [DOI] [PubMed] [Google Scholar]
- Ando T, Rasmussen K, Nyhan WL, and Hull D (1972). 3-hydroxypropionate: significance of -oxidation of propionate in patients with propionic acidemia and methylmalonic acidemia. Proc. Natl. Acad. Sci. USA 69, 2807–2811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arda HE, Taubert S, Conine C, Tsuda B, Van Gilst MR, Sequerra R, Doucette-Stam L, Yamamoto KR, and Walhout AJM (2010). Functional modularity of nuclear hormone receptors in a Caenorhabditis elegans metabolic gene regulatory network. Mol. Syst. Biol. 6, 367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babicki S, Arndt D, Marcu A, Liang Y, Grant JR, Maciejewski A, and Wishart DS (2016). Heatmapper: web-enabled heat mapping for all. Nucleic Acids Res. 44, W147–W153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner S (1974). The genetics of Caenorhabditis elegans. Genetics 77, 71–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deodato F, Boenzi S, Santorelli FM, and Dionisi-Vici C (2006). Methylmalonic and propionic aciduria. Am. J. Med. Genet. C. Semin. Med. Genet. 142C, 104–112. [DOI] [PubMed] [Google Scholar]
- Dupuy D, Bertin N, Hidalgo CA, Venkatesan K, Tu D, Lee D, Rosenberg J, Svrzikapa N, Blanc A, Carnec A, et al. (2007). Genome-scale analysis ofin vivo spatiotemporal promoter activity in Caenorhabditis elegans. Nat. Biotechnol. 25, 663–668. [DOI] [PubMed] [Google Scholar]
- Evans RM, and Mangelsdorf DJ (2014). Nuclear receptors, RXR, and the Big Bang. Cell 157, 255–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frøkjaer-Jensen C, Davis MW, Sarov M, Taylor J, Flibotte S, LaBella M, Pozniakovsky A, Moerman DG, and Jorgensen EM (2014). Random and targeted transgene insertion in Caenorhabditis elegans using a modified Mos1 transposon. Nat. Methods 11, 529–534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuxman Bass JI, Pons C, Kozlowski L, Reece-Hoyes JS, Shrestha S, Holdorf AD, Mori A, Myers CL, and Walhout AJM (2016). A gene-centered C. elegans protein-DNA interaction network provides a framework for functional predictions. Mol. Syst. Biol. 12, 884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamath RS, Fraser AG, Dong Y, Poulin G, Durbin R, Gotta M, Kanapin A, Le Bot N, Moreno S, Sohrmann M, et al. (2003). Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature 421,231–237. [DOI] [PubMed] [Google Scholar]
- Leng N, Dawson JA, Thomson JA, Ruotti V, Rissman AI, Smits BM, Haag JD, Gould MN, Stewart RM, and Kendziorski C (2013). EBSeq: an empirical Bayes hierarchical model for inference in RNA-seq experiments. Bioinformatics 29, 1035–1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung CK, Deonarine A, Strange K, and Choe KP (2011). High-throughput screening and biosensing with fluorescent C. elegans strains. J. Vis. Exp. 19, 2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacNeil LT, Watson E, Arda HE, Zhu LJ, and Walhout AJM (2013). Diet-induced developmental acceleration independent of TOR and insulin in C. elegans. Cell 153, 240–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacNeil LT, Pons C, Arda HE, Giese GE, Myers CL, and Walhout AJM (2015). Transcription factor activity mapping of a tissue-specific in vivo gene regulatory network. Cell Syst. 1, 152–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangan S, and Alon U (2003). Structure and function of the feed-forward loop network motif. Proc. Natl. Acad. Sci. USA 100, 11980–11985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGhee JD, Sleumer MC, Bilenky M, Wong K, McKay SJ, Goszczynski B, Tian H, Krich ND, Khattra J, Holt RA, et al. (2007). The ELT-2 GATA-factor and the global regulation of transcription in the C. elegans intestine. Dev. Biol. 302, 627–645. [DOI] [PubMed] [Google Scholar]
- Peters H, Ferdinandusse S, Ruiter JP, Wanders RJ, Boneh A, and Pitt J (2015). Metabolite studies in HIBCH and ECHS1 defects: implications for screening. Mol. Genet. Metab. 115, 168–173. [DOI] [PubMed] [Google Scholar]
- Reece-Hoyes JS, Pons C, Diallo A, Mori A, Shrestha S, Kadreppa S, Nelson J, Diprima S, Dricot A, Lajoie BR, et al. (2013). Extensive rewiring and complex evolutionary dynamics in a C. elegans multiparameter transcription factor network. Mol. Cell 51, 116–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rual J-F, Ceron J, Koreth J, Hao T, Nicot A-S, Hirozane-Kishikawa T, Vandenhaute J, Orkin SH, Hill DE, van den Heuvel S, and Vidal M (2004). Toward improving Caenorhabditis elegans phenome mapping with an ORFeome-based RNAi library. Genome Res. 14, 2162–2168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson E, MacNeil LT, Arda HE, Zhu LJ, and Walhout AJM (2013). Integration of metabolic and gene regulatory networks modulates the C. elegans dietary response. Cell 153, 253–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson E, MacNeil LT, Ritter AD, Yilmaz LS, Rosebrock AP, Caudy AA, and Walhout AJM (2014). Interspecies systems biology uncovers metabolites affecting C. elegans gene expression and life history traits. Cell 156, 759–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson E, Olin-Sandoval V, Hoy MJ, Li C-H, Louisse T, Yao V, Mori A, Holdorf AD, Troyanskaya OG, Ralser M, and Walhout AJ (2016). Metabolic network rewiring of propionate flux compensates vitamin B12 deficiency in C. elegans. eLife 5, e17670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yilmaz LS, and Walhout AJ (2016). A Caenorhabditis elegans genome-scale metabolic network model. Cell Syst. 2, 297–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The RNA-sequencing data files were deposited in the NCBI Gene Expression Omnibus (GEO) under accession number GEO: GSE123507.