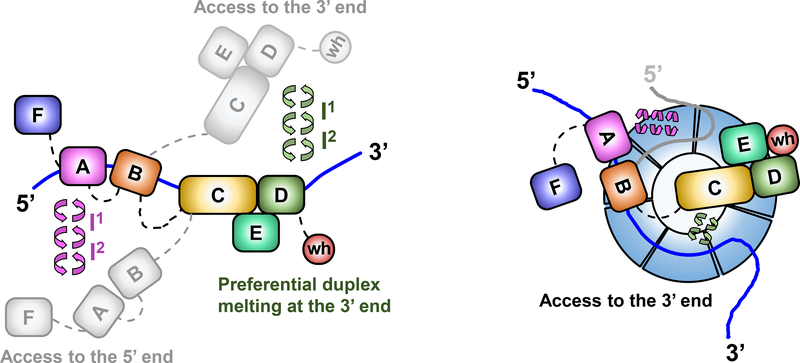
Figure 4. Dynamics of RPA DBDs and modulation by Rad52.
a) Sequential and directional arrangement of the DBDs allows RPA to occlude 20–30 nt of ssDNA (∼20 nt under our experimental conditions; Supplemental Fig 1). When RPA is in a stoichiometric complex with ssDNA, or when the ssDNA is in excess, the individual DBDs of RPA exist in a variety of distinct dynamic conformational DNA bound states. Such conformational flexibility allows access to either the 5′ or the 3′ segment of the DNA to other proteins that function in downstream processes. The circular arrows represent the transitions between multiple fluorescence states we observe in the single molecule experiments and which are implied by the bulk stopped flow experiments. Note that while we illustrate the changes in the conformation of the RPA-ssDNA complex as movement of the DBDs, the same microscopically bound states may arise from ssDNA dissociating and moving away from the respective DBDs. b) The DBDs are also selectively modulated by RPA-interacting proteins (RIPs) such as Rad52. In this case, only the DNA binding dynamics of DBD-D, and possibly the trimerization core, is influenced by Rad52. In the ternary RPA-ssDNA-Rad52 complex, the ssDNA is shared by RPA and Rad52, which also interact with one another. The ability of the DBD-D and other RPA elements contacting the ssDNA near the 3′ end of the occluded sequence is constrained. Such selective DBD modulation may promote loading of Rad51 onto the 3′ end of the ssDNA during homologous recombination.