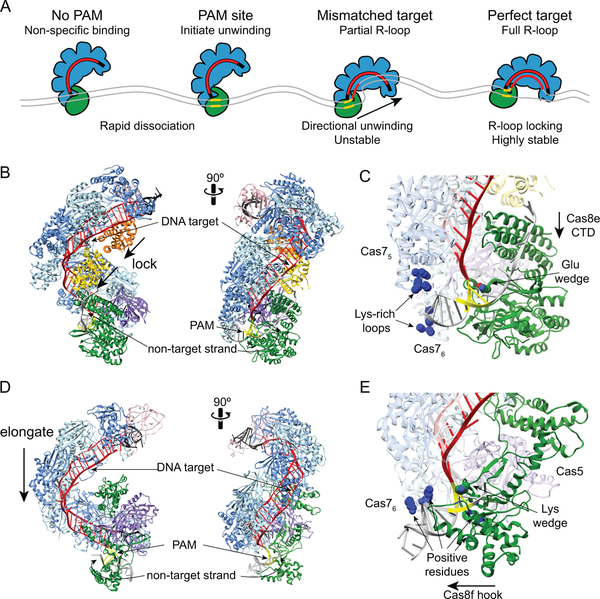
Figure 7.
Target search and R-loop formation by Cascade and Csy complexes. A. Overview of target search by E. coli Cascade. Non-specific interactions at sites with no PAM or PAMs adjacent to non-matching sequences result in rapid dissociation. At PAM sites, Cascade can initiate unwinding to enable crRNA strand invasion. At partially matching target, R-loops can form directionally away from the PAM until a mismatch is encountered. Depending on the location and thermodynamic barrier of the mismatch, the R-loop can stall, continue to form or Cascade can dissociate. At a perfect target, complete R-loop formation leads to Cascade locking and a highly stable interaction. B. Structure of E. coli Cascade bound to dsDNA (PDB ID 5H9F, colored as in Figure 6) (127). The Cas11 subunits move down along the belly during R-loop locking. The non-target strand is stabilized through interactions with Cas8e. C. Close-up of Cas8e-PAM and Cascade-dsDNA interactions. A glutamate residue acts as a wedge to facilitate DNA unwinding. The PAM is read through minor-groove interactions with Cas8e. Lys-rich loops in Cas75 and Cas76 subunits act as an electrostatic vise to hold the dsDNA during unwinding. The Cas8e CTD four-helix bundle moves toward the N-terminal domain during R-loop locking. D. Structure of P. aeruginosa Csy complex bound to dsDNA (PDB ID 6B44, colored as in Figure 6) (59). The Cas7 backbone elongates by ~20 Å upon DNA binding. E. Close-up of Cas8f-PAM and Csy-dsDNA interactions. A lysine residue acts as a wedge to facilitate DNA unwinding. The PAM is read through minor-groove interactions with Cas8f. Several positive residues in Cas76, Cas8f and Cas5 stabilize interactions with the dsDNA. The Cas8f hook elongates to accommodate the dsDNA.