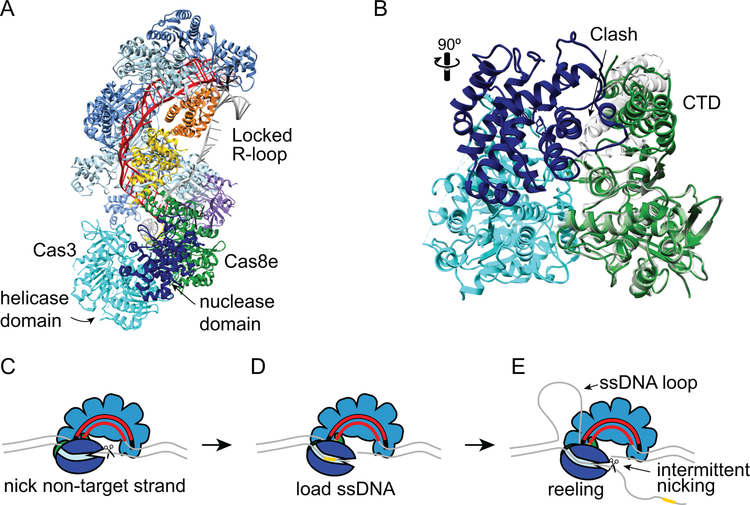
Figure 8.
Target degradation by Cas3 during CRISPR interference. A. Structure of type I-E T. fusca Cascade-dsDNA following Cas3 recruitment (PDB ID 6C66) (33). Cascade and DNA are colored as in Figures 6 and 7, Cas3 is shown in cyan (helicase and C-terminal domains) and dark blue (HD nuclease domain). Cas3 senses the conformation of Cas8e following R-loop locking. Comparison of Cas8e structure in the locked (PDB ID 6C66, green) and unlocked (PDB ID 5U07, gray) conformations (33, 128). Prior to R-loop locking and movement of the CTD, a loop and helix of the Cas8e CTD would cause steric clashes with the Cas3 nuclease domain. C-E. Schematic of E. coli Cas3 DNA reeling and degradation. C. Following its recruitment to the R-loop, Cas3 nicks the non-target strand. D. The nicked strand is loaded into the Cas3 helicase domain, stabilizing the Cas3-Cascade interaction. E. The Cas3 helicase domain reels the non-target strand, while the target strand is extruded and forms a ssDNA loop. The HD nuclease domain can nick the non-target strand intermittently, creating products that could serve as prespacer substrates for Cas1-Cas2 (Figure 3E), or it can release the DNA and repeat the reeling motion. In type I-F systems, two Cas2/3 molecules may be recruited to the Csy-complex, allowing for bidirectional degradation of both strands of the DNA.