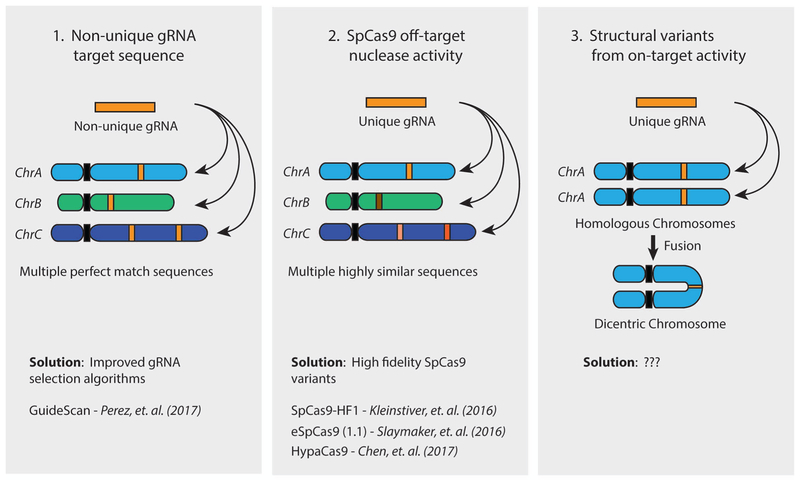
Figure 3 – Sources of SpCas9 off-target activity.
The activity of SpCas9 in human cells can produce off-target consequences through three mechanisms, based on the gRNA being used. (1) If the 20nt gRNA target sequence is not unique in the human genome, SpCas9 will induce a DNA double strand break (DSB) at each locus containing a perfect match, resulting in the formation of non-targeted indels or structural rearrangements. This can be avoided through the use of gRNA selection algorithms specifically designed to avoid non-unique sequences, such as GuideScan. (2) SpCas9 can also induce DSBs at highly similar sequences containing 3 or fewer mismatches to the gRNA. These off-target breaks can result in non-targeted indels and structural rearrangements either alone or in combination with on-target SpCas9 activity. The propensity for SpCas9 to cut at mismatched target sites can be reduced through the use of reported high-fidelity SpCas9 variants, including SpCas9-HF1, eSpCas9, and HypaCas9. (3) Even completely on-target activity from a single gRNA has the potential to introduce off-target structural rearrangements through simultaneous cutting of the same locus on homologous chromosomes or sister chromatids. Chromosome head-to-head fusion is one potential outcome of this event, resulting in the formation of dicentric chromosome. No solution for this type of off-target event has yet been proposed.