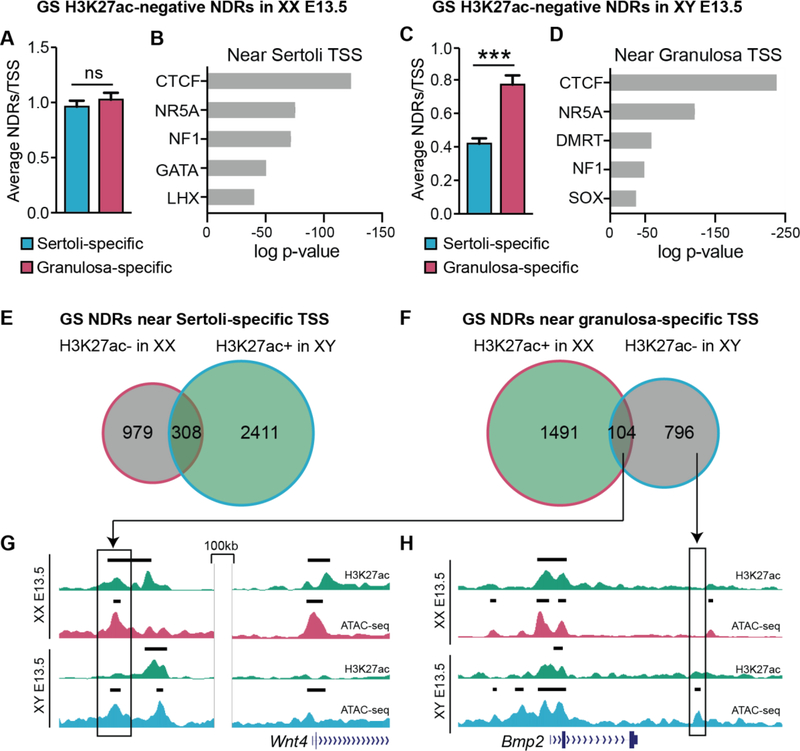
Figure 3. Gonad-specific H3K27ac-negative NDRs are potential silencers of the alternate fate.
Analysis of gonad-specific (GS) H3K27ac-negative NDRs in granulosa cells (A&B), and GS H3K27ac-negative NDRs in Sertoli cells (C&D). A&C) Graphs showing the average number of NDRs associated to the nearest TSS as determined by HOMER. NDRs were associated to the nearest TSS of genes that become granulosa cell-specific (pink) and Sertoli cell-specific (blue). Error bars represent the standard error of the mean (SEM) and *** represents p<0.0001 as determined by a student’s t test. B&D) Top five motifs identified by TF binding motif analysis of protein families with members expressed in the gonad ranked in order of significance (p<e10, additional motifs in Table S1). E&F) Venn diagrams of H3K27ac-negative (grey) and H3K27ac-positive (green) NDRs in granulosa cells (pink outline) and Sertoli cells (blue outline), near Sertoli-specific TSS (E) and granulosa-specific TSS (F). G&H) Genome Browser tracks of ATAC-seq in granulosa (dark pink) and Sertoli (dark blue), and corresponding ChIP-seq for H3K27ac (green). Bold black lines above tracks represent significant enrichment compared to flanking regions as determined by HOMER. An NDR near the granulosa-promoting gene Wnt4 that is H3K27ac-positive in granulosa cells and H3K27ac-negative in Sertoli cells (G) and an NDR near the granulosa-promoting gene Bmp2 that is H3K27ac-negative in Sertoli cells and is not shared in granulosa cells (H) are shown.