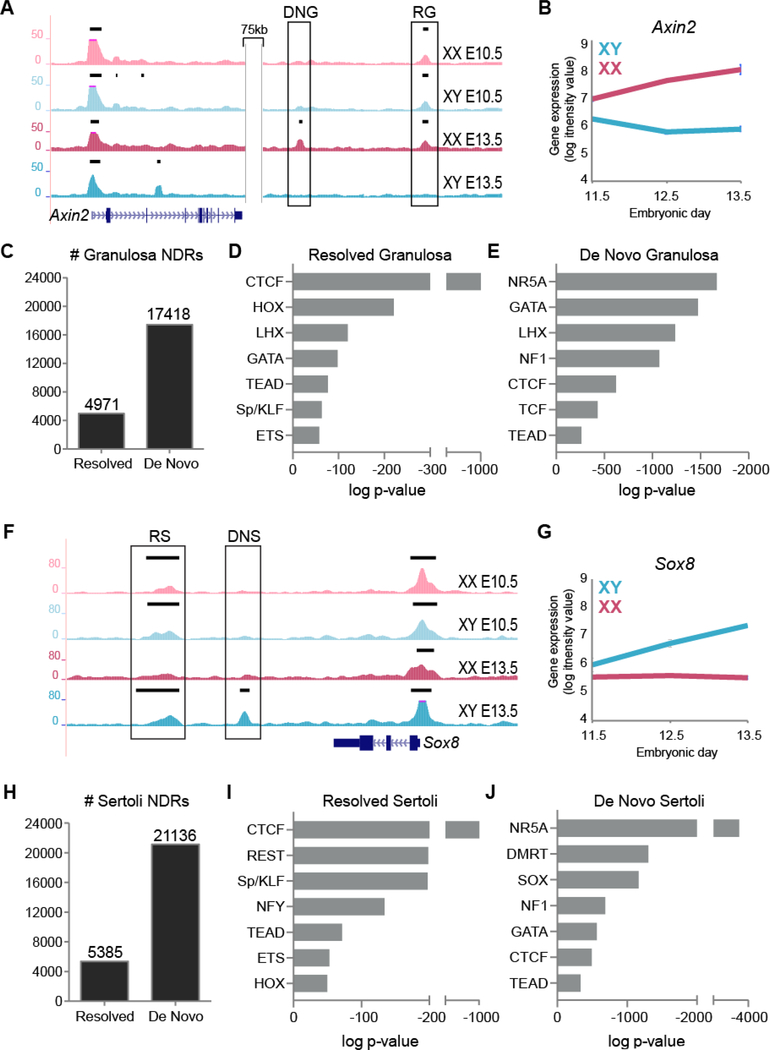
Figure 4. Gonad-specific NDRs are either retained from the progenitor state or arise de novo.
A) Genome Browser tracks of ATAC-seq in XX E10.5 (light pink), XY E10.5 (light blue), granulosa (dark pink) and Sertoli (dark blue) purified cells of the genomic location surrounding Axin2. Bold black lines above tracks represent significant enrichment compared to flanking regions as determined by HOMER. A resolved granulosa (RG) and a de novo granulosa (DNG) NDR (boxed) lie downstream of Axin2. B) Axin2 gene expression profile in purified XX (pink) and XY (blue) supporting cells throughout sex determination (E11.5-E13.5) (Jameson et al., 2012b). C) Number of resolved and de novo NDRs in granulosa cells. D and E) Top motifs of families with members expressed in the gonad identified by TF binding motif analysis in resolved and de novo NDRs in granulosa cells ranked in order of significance (additional motifs in Table S1). F) Genome Browser tracks of ATAC-seq in XX E10.5 (light pink), XY E10.5 (light blue), granulosa (dark pink) and Sertoli (dark blue) purified cells of the genomic location surrounding Sox8. Bold black lines above tracks represent significant enrichment compared to flanking regions as determined by HOMER. A resolved Sertoli (RS) and a de novo Sertoli (DNS) NDR (boxed) lie downstream of Sox8. G) Sox8 gene expression profile in purified XX (pink) and XY (blue) supporting cells throughout sex determination (E11.5-E13.5) (Jameson et al., 2012b). H) Number of resolved and de novo NDRs in Sertoli cells. I and J) Top motif families (with members expressed in the gonad) identified by TF binding motif analysis in resolved and de novo NDRs in Sertoli cells ranked in order of significance (additional motifs in Table S1).